Fig. 2:
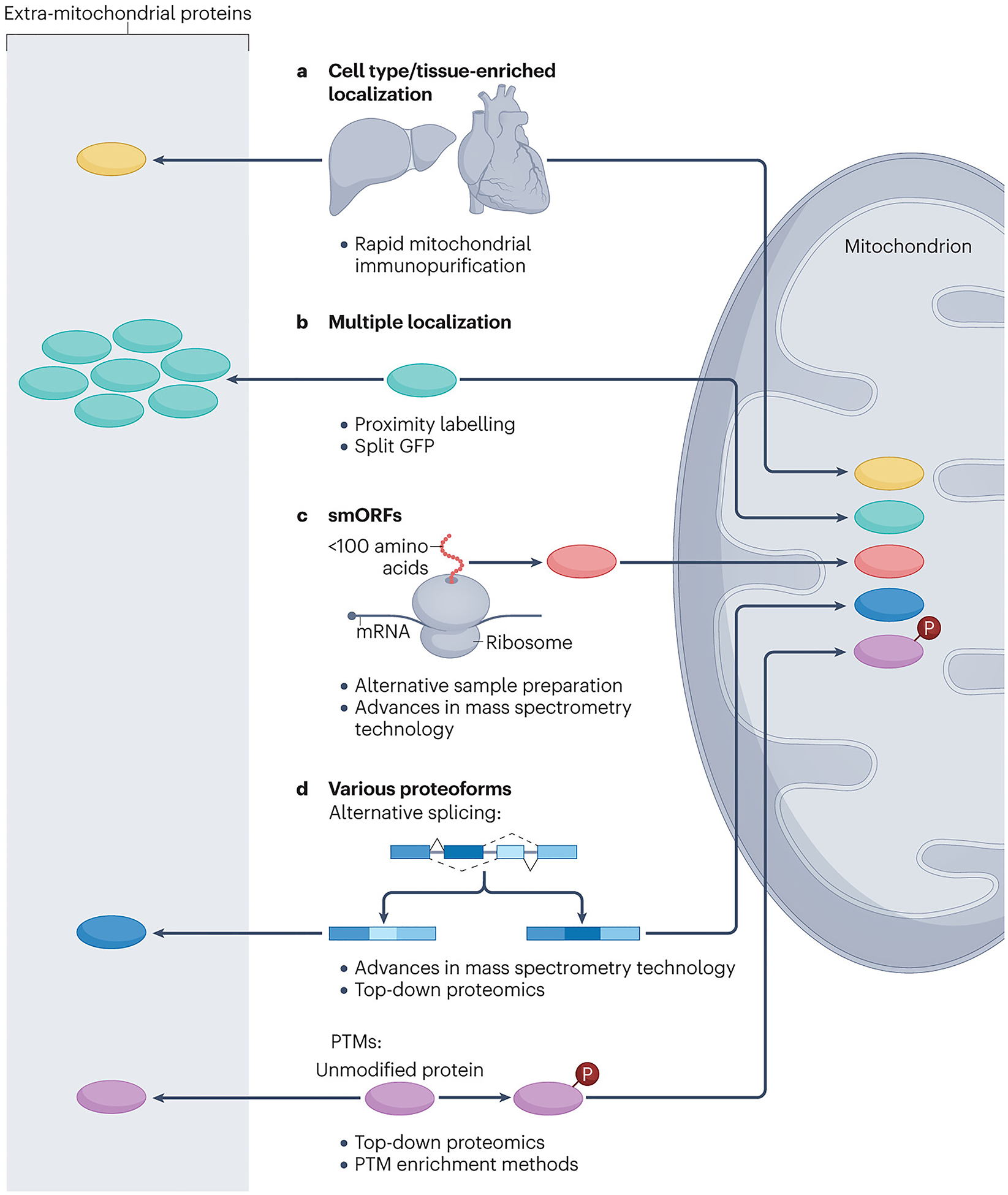
Potential sources of new mitochondrial proteins.
New mitochondrial proteins can derive from different sources. a, Protein expression is differential across different tissues and cell types; therefore, the expression of mitochondrial proteins is variable too. To capture mitochondrial proteins in specific tissues, epitope-tagged outer mitochondrial membrane proteins can be utilized as handles for immunocapture. b, Proteins with multiple localizations (including mitochondrial localization) cannot be found by means of subtractive proteomics if the protein of interest is low in abundance in the mitochondrial fraction. These proteins can be identified through proximity labelling, which allows specific tagging of proteins that reside in different mitochondrial subcompartments. c, Small open reading frame (smORF) proteins can be identified as mitochondrial proteome members through the use of alternative digestion strategies and by mass spectrometry search strategies that include peptides corresponding to predicted smORFs to address the unique features of these proteins. d, Identifying the variety of proteoforms can be facilitated by adjusting mass spectrometry search strategies to reflect the different peptide compositions of protein isoforms and by enrichment techniques for post-translational modifications (PTMs), for example, phosphorylated peptides (Box 2).
