Fig. 4:
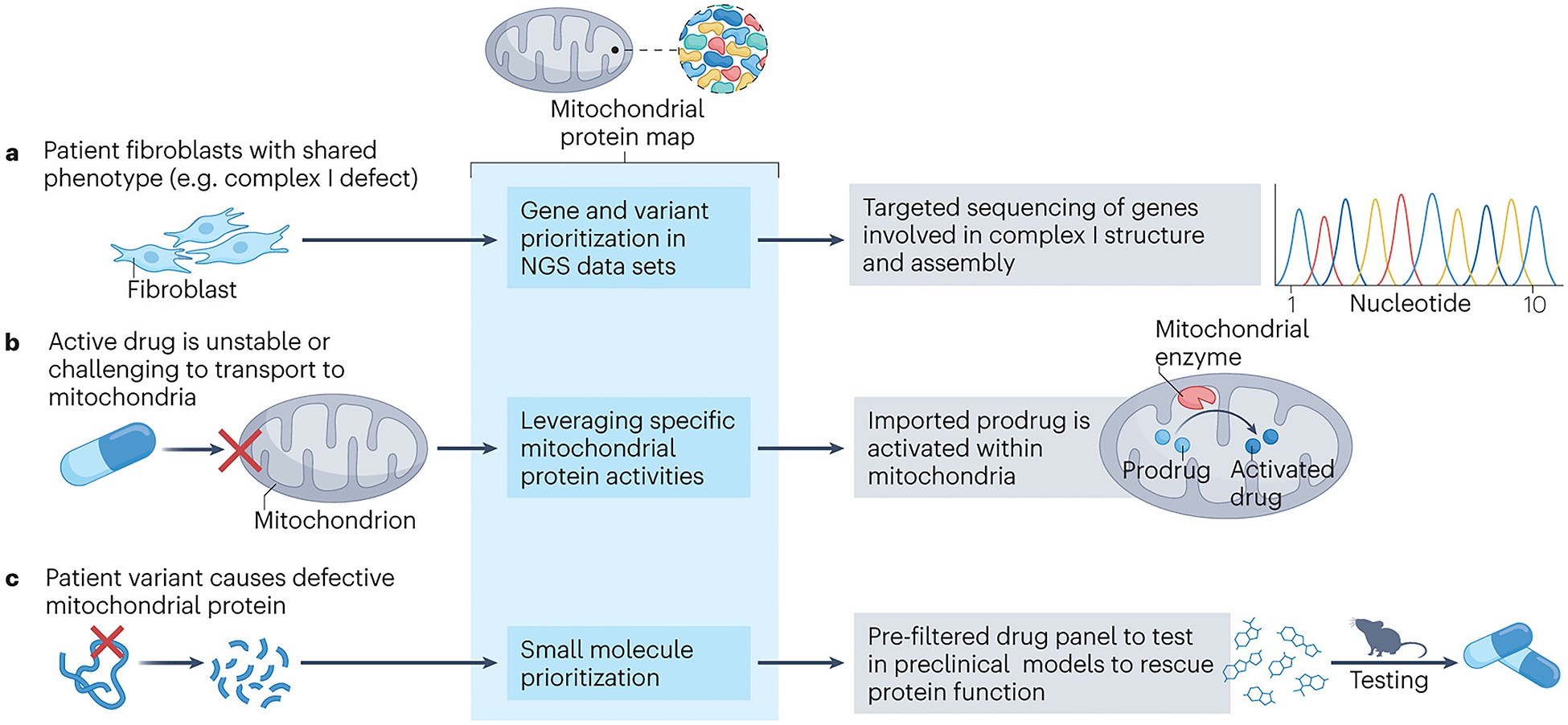
Translational utility of the mitochondrial protein map.
The mitochondrial protein map can prove useful in multiple translational scenarios of which three examples are depicted. a, The diagnostic challenge posed by primary mitochondrial diseases can be effectively addressed through an elegant gene prioritization approach facilitated by the mitochondrial protein map. By comparing phenotypic traits in diagnostic samples with protein function information in the mitochondrial protein map, a specific group of genes can be triaged for diagnostic sequencing. For instance, when faced with patient samples displaying biochemical complex I deficiency, employing a prioritized variant search within the genes associated with complex I function and assembly, as annotated in the mitochondrial protein map, could expedite genetic diagnosis. b, Drug targeting of mitochondrial proteins is challenging due to insufficient transport of specific substances to their intended mitochondrial targets or premature drug degradation. However, this challenge can be addressed by devising prodrugs that exhibit both stability and the ability to traverse mitochondrial membranes. Once these prodrugs reach the mitochondrial matrix, they can undergo processing, such as cleavage, which activates them and enables them to exert their desired effect on the target mitochondrial protein. The rational design of prodrug processing within mitochondria can be facilitated by the comprehensive information provided by the mitochondrial protein map, which offers detailed insights into the resident mitochondrial enzymes that can be harnessed for effective prodrug processing. c, Primary mitochondrial diseases arise from variants that directly impact the optimal functionality of mitochondrial proteins. In the specific case shown here, the variant leads to a disruption in protein stability, presenting a potential avenue for intervention through protein stabilization by small molecules. To optimize this approach, we recommend that the future mitochondrial protein map incorporates a comprehensive library of small molecules, characterized for their specific interactions with mitochondrial proteins. Leveraging this information, small molecules can be prioritized for testing purposes, focusing on those with promising potential to exert beneficial effects on the deficient protein. NGS, next-generation sequencing.
