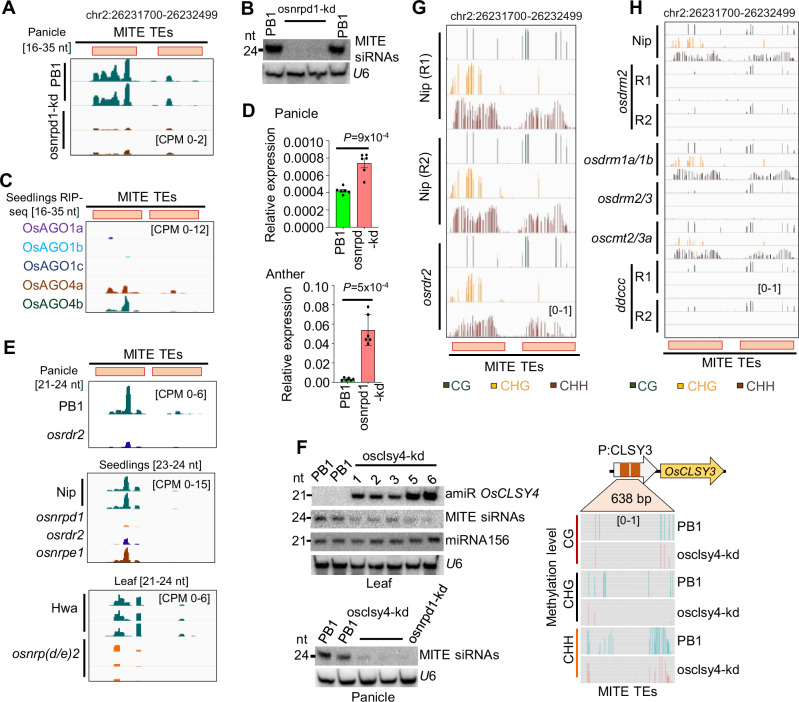
Fig. 2. DNA methylation in OsCLSY3 promoter is regulated by RdDM.
A Integrated Genomics Viewer (IGV) screenshots showing OsCLSY3 promoter derived sRNA levels (16–35 nt) in osnrpd1-kd panicle. B Northern blot showing sRNA levels in osnrpd1-kd panicle. C IGV screenshots showing enrichment of OsCLSY3 promoter derived sRNAs (16–35 nt) in RIP of different AGOs in rice (GSE18250 and GSE20748). D Barplots showing OsCLSY3 level in osnrpd1-kd panicles (top) and osnrpd1-kd anther (bottom). OsActin served as internal control. Data represents mean ± SE of 2 independent biological replicates with 3 technical replicates each (n = 6). Comparisons were made with paired two-tailed Student’s t-tests (P < 0.05 considered as significant). E IGV screenshots showing status of MITE derived sRNAs among RdDM mutants of rice (GSE130166, GSE158709, Nip—Japonica cultivar Nipponbare) (PRJNA758109, Hwa—Japonica cultivar Hwayoung). F Northern blots showing MITE derived sRNA level in osclsy4-kd and osnrpd1-kd (left panels) and analysis of DNA methylation in OsCLSY3 promoter in osclsy4-kd leaf by BS-PCR (right panel). U6 served as loading control. G IGV screenshots showing methylation status of 800 bp MITE TE region in OsCLSY3 promoter in osrdr2 seedlings (GSE130168). H IGV screenshot showing methylation status in leaves of ddccc mutant (GSE138705). ddccc is osdrm2/osdrm1/oscmt2a/oscmt2b/oscmt3 (Nip—Japonica cultivar Nipponbare). Source data are provided as a Source Data file. (B), (D) and (F) were repeated at least twice with consistent results.