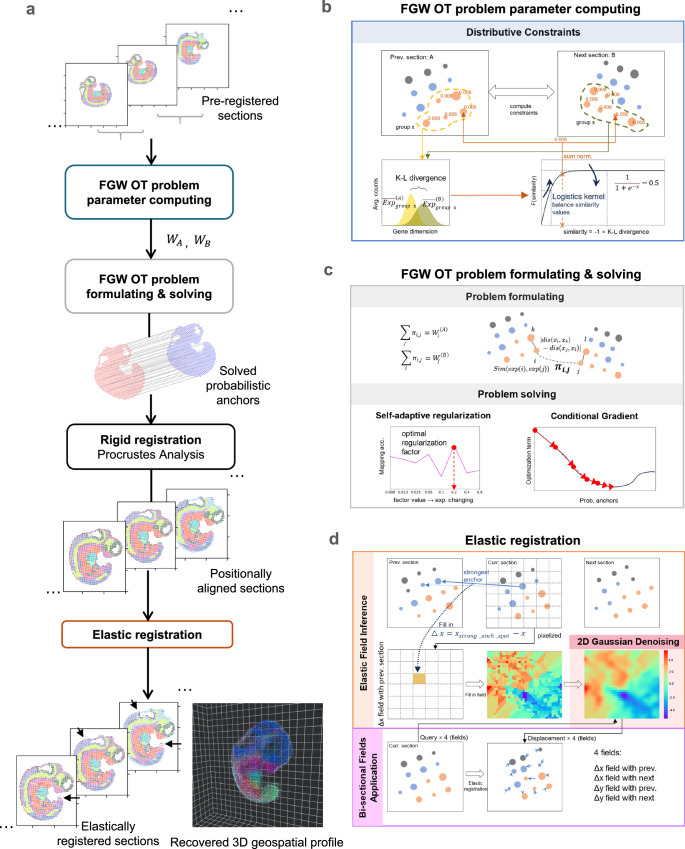
Fig. 1. Three-Dimensional (3D) Spatial Transcriptomics (ST) Geospatial profile recovery with ST-GEARS.
a The automatic pipeline of ST-GEARS which recovers ST-GEARS 3D in vivo spatial information by ordered steps including Fused Gromov Wasserstein (FGW) Optimal Transport (OT) problem parameter computing, problem formulating and solving which outputs probabilistic anchors across sections, rigid registration through Procrustes Analysis which solves optimal positional alignment using the anchors, and finally elastic registration. The input of the method is Unique molecular identifier (UMI) counts and location of each spot measured by ST technology, along with their annotations or cross-section clustering result. And the output of the method is recovered 3D in vivo spatial information of the experimented tissue, or sample. b FGW OT problem parameter computing, which assigns nonuniform weights to spots in preparation for future problem formulating, based on cross-sectional similarity of annotation types or clusters. c FGW OT problem formulating, whose setting aims to solve probabilistic anchors joining spots with highest in vivo proximity, through optimizing the combination of gene expression and structural similarity34. FGW OT problem solving, which is implemented based on Conditional Gradient (CG) method, leading to retrieved probabilistic anchors. d Elastic registration, which utilizes the anchors again to compute and denoise distortion fields which guides the elimination of distortions, then applies the fields bi-sectionally to positionally aligned sections, leading to the recovered 3D in vivo spatial information.