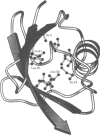
Abstract
The solid-phase chemical synthesis of ubiquitin produced a molecule with physicochemical properties similar to those of the natural protein. We have grown crystals of this synthetic ubiquitin and performed an X-ray analysis at 1.8 A resolution in order to compare the synthetic protein with the known natural structure. The crystals were isomorphous with those of the natural protein, the R-factor between them being 7.1%. Difference Fourier analysis shows that the synthetic and natural structures are indistinguishable. The co-ordinates of the natural ubiquitin (1UBQ) were used as the starting point for restrained least-squares refinement (TNT program) against the synthetic X-ray data. The refinement converged to R = 16.5% and the resulting model did not change when refined against natural ubiquitin X-ray data (R = 18.7%). From both the refinement and featureless difference Fourier synthesis, we conclude that the synthetic and natural protein structures are identical. A short discussion about the uses of proteins with 'non-standard' amino acid residues is included.
Full text
PDF
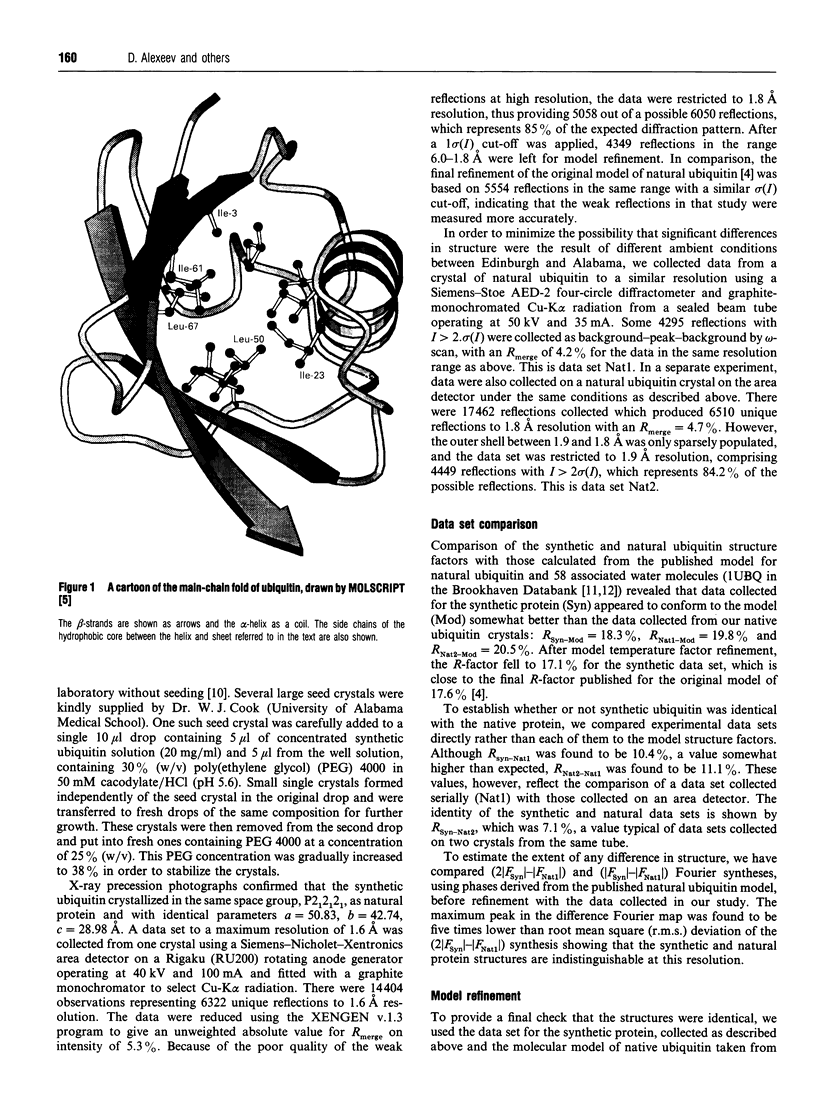
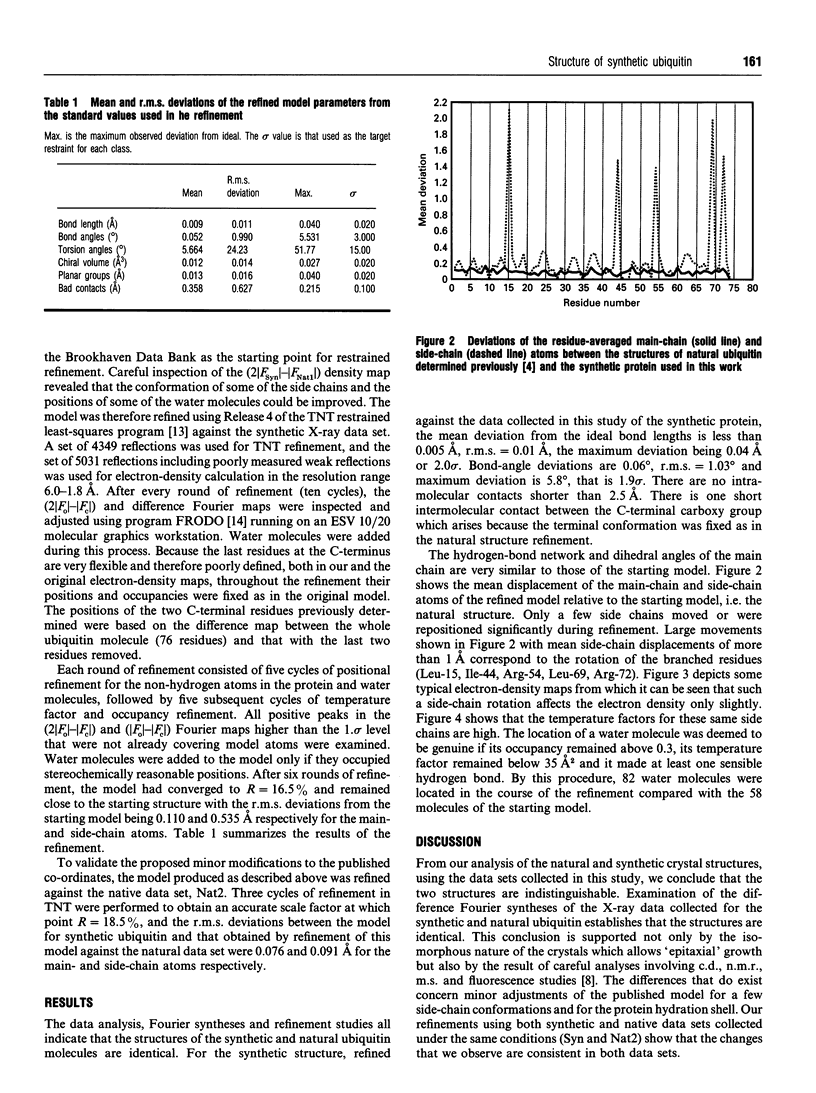
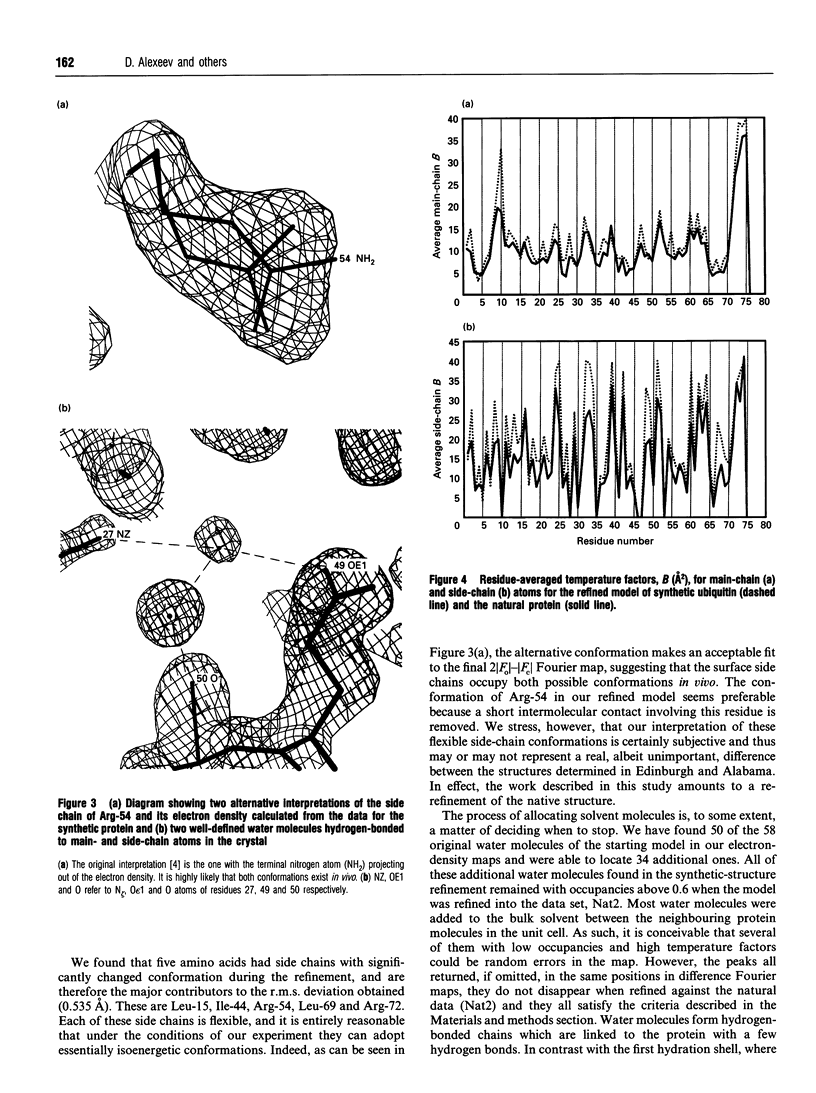

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bernstein F. C., Koetzle T. F., Williams G. J., Meyer E. F., Jr, Brice M. D., Rodgers J. R., Kennard O., Shimanouchi T., Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977 May 25;112(3):535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- Briggs M. S., Roder H. Early hydrogen-bonding events in the folding reaction of ubiquitin. Proc Natl Acad Sci U S A. 1992 Mar 15;89(6):2017–2021. doi: 10.1073/pnas.89.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham B. C., Wells J. A. Improvement in the alkaline stability of subtilisin using an efficient random mutagenesis and screening procedure. Protein Eng. 1987 Aug-Sep;1(4):319–325. doi: 10.1093/protein/1.4.319. [DOI] [PubMed] [Google Scholar]
- Ellis R. J., van der Vies S. M. Molecular chaperones. Annu Rev Biochem. 1991;60:321–347. doi: 10.1146/annurev.bi.60.070191.001541. [DOI] [PubMed] [Google Scholar]
- Finley D., Bartel B., Varshavsky A. The tails of ubiquitin precursors are ribosomal proteins whose fusion to ubiquitin facilitates ribosome biogenesis. Nature. 1989 Mar 30;338(6214):394–401. doi: 10.1038/338394a0. [DOI] [PubMed] [Google Scholar]
- Freedman R. B., Bulleid N. J., Hawkins H. C., Paver J. L. Role of protein disulphide-isomerase in the expression of native proteins. Biochem Soc Symp. 1989;55:167–192. [PubMed] [Google Scholar]
- Hershko A., Ciechanover A. The ubiquitin system for protein degradation. Annu Rev Biochem. 1992;61:761–807. doi: 10.1146/annurev.bi.61.070192.003553. [DOI] [PubMed] [Google Scholar]
- Hurley J. H., Baase W. A., Matthews B. W. Design and structural analysis of alternative hydrophobic core packing arrangements in bacteriophage T4 lysozyme. J Mol Biol. 1992 Apr 20;224(4):1143–1159. doi: 10.1016/0022-2836(92)90475-y. [DOI] [PubMed] [Google Scholar]
- Ramage R., Green J., Muir T. W., Ogunjobi O. M., Love S., Shaw K. Synthetic, structural and biological studies of the ubiquitin system: the total chemical synthesis of ubiquitin. Biochem J. 1994 Apr 1;299(Pt 1):151–158. doi: 10.1042/bj2990151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thillet J., Absil J., Stone S. R., Pictet R. Site-directed mutagenesis of mouse dihydrofolate reductase. Mutants with increased resistance to methotrexate and trimethoprim. J Biol Chem. 1988 Sep 5;263(25):12500–12508. [PubMed] [Google Scholar]
- Vijay-Kumar S., Bugg C. E., Cook W. J. Structure of ubiquitin refined at 1.8 A resolution. J Mol Biol. 1987 Apr 5;194(3):531–544. doi: 10.1016/0022-2836(87)90679-6. [DOI] [PubMed] [Google Scholar]