Figure 2. Two-Hit Genetic Event in TNK2 and Protein Modeling of the p.M171T Variant.
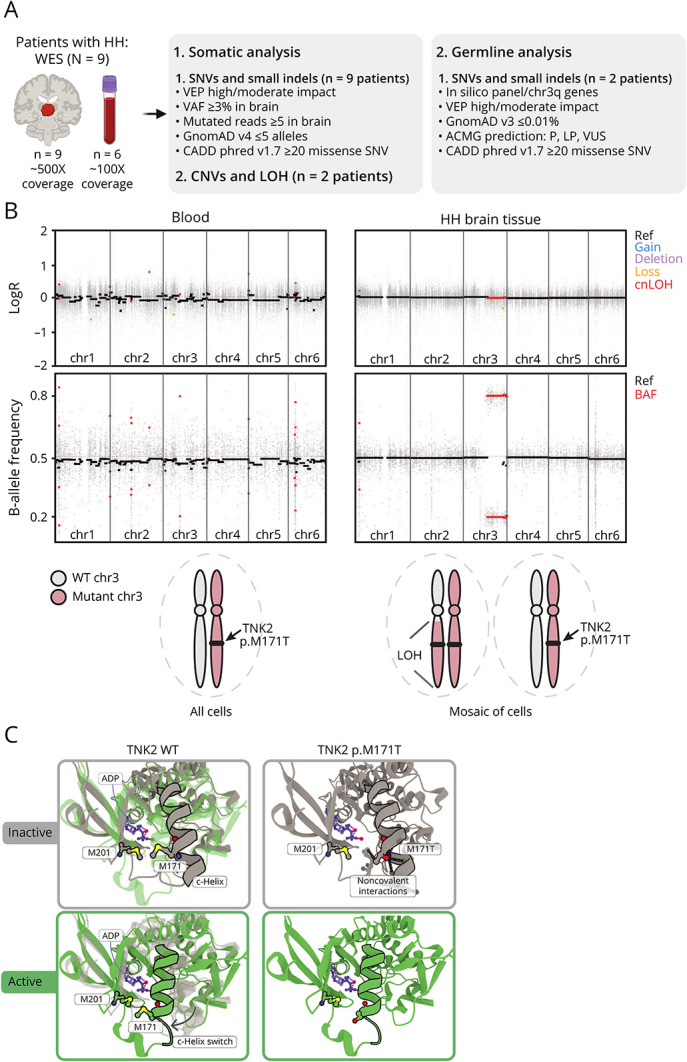
(A) Whole-exome sequencing (WES) and bioinformatic analysis workflow. (B) Top: somatic loss-of-heterozygosity (LOH) on chromosome 3q (chr3q29) detected in HH brain tissue (but not in the blood) from patient ICM_212 (B-allele frequency distribution). The absence of chromosome gain or loss (LogR ratio) on chr3q indicates copy-neutral LOH (cnLOH). Bottom: schematic of wild-type (WT) and mutant TNK2 loci in blood and HH brain cells. (C) In silico 3D modeling of the inactive (grey) and active (green) states of the TNK2 kinase domain. The p.M171 residue lies within a C-helix, whose inward switch is required for the activation of kinase domains.9 In the WT configuration, the M171 points toward the M201, displaying hydrophobic interactions (pink halo) that are lost in the mutant TNK2. The p.M171T is predicted to destabilize the inactive state of TNK2. ACMG = American College of Medical Genetics and Genomics; ADP = adenosine diphosphate; BAF, B-allele frequency; CNVs = copy number variants; HH = hypothalamic hamartoma; indels = insertions/deletions; LOH = loss-of-heterozygosity; LP = likely pathogenic; P = pathogenic; Ref, reference; SNV = single-nucleotide variant; VAF = variant allele frequency; VEP = variant effect predictor (Ensembl); VUS = variant of unknown significance.
