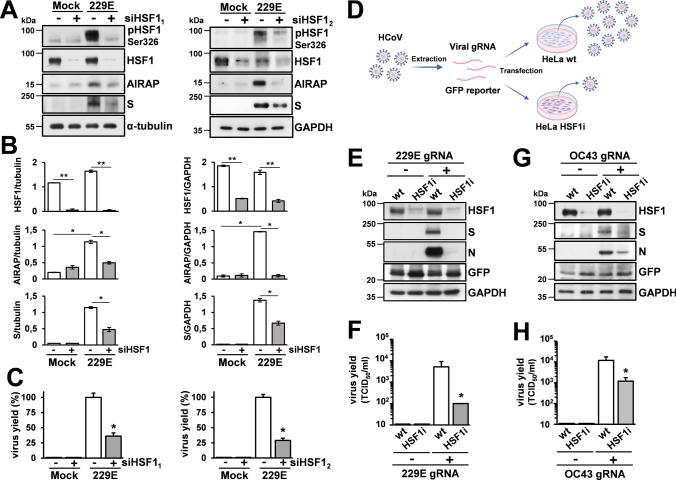
Fig. 5.
HSF1 is required for efficient HCoV replication. A Immunoblot of pHSF1-Ser326, HSF1, AIRAP, viral spike (S), α-tubulin and GAPDH protein levels in MRC-5 cells transiently transfected with two different HSF1-siRNAs [siHSF11 (left) and siHSF12 (right); +] or scramble-RNA (−) for 48 h, and infected with HCoV-229E (0.1 TCID50/cell) or mock infected (Mock) for 24 h. B The relative amount of total HSF1, AIRAP and viral S protein, normalized to the loading control in the same sample, were determined by densitometric analysis using ImageJ software. Error bars indicate means ± S.D. (n = 3). C In parallel, virus yield from supernatants of infected cells was determined at 24 h p.i. by TCID50 infectivity assay. Data, expressed as percentage of control, represent the mean ± S.D. (n = 3). *p < 0.05, **p < 0.01; Student’s t-test (B, C). D Schematic representation of HCoV genomic RNA transfection assays. E–H Wild-type (wt) or stably HSF1-silenced (HSF1i) HeLa cells were co-transfected with HCoV-229E (229E gRNA) or HCoV-OC43 (OC43 gRNA) genomic RNA and the pCMV-GFP vector for 4 h. After 48 h (229E) or 72 h (OC43), levels of HSF1, viral spike (S) and nucleocapsid (N), GFP and GAPDH proteins were analyzed by IB (E, G). In parallel, virus yield in the supernatant of transfected cells was determined by TCID50 infectivity assay (F, H). Data, expressed as TCID50/ml, represent the mean ± S.D. (n = 3). *p < 0.05; Student’s t-test