Abstract
Our genome is exposed to a wide variety of DNA-damaging agents. If left unrepaired, this damage can be converted into mutations that promote carcinogenesis or the development of genetically inherited diseases. As a result, researchers and clinicians require tools that can detect DNA damage and mutations with exceptional sensitivity. In this study, we describe a massively parallel sequencing tool termed Mutation And DNA Damage Detection-seq (MADDD-seq) that is capable of detecting O6-methyl guanine lesions and mutations simultaneously, with a single assay. To illustrate the dual capabilities of MADDD-seq, we treated WT and DNA repair deficient yeast cells with the DNA-damaging agent MNNG and tracked DNA lesions and mutations over a 24-h time period. This approach allowed us to identify thousands of DNA adducts and mutations in a single sequencing run and gain deep insight into the kinetics of DNA repair and mutagenesis.
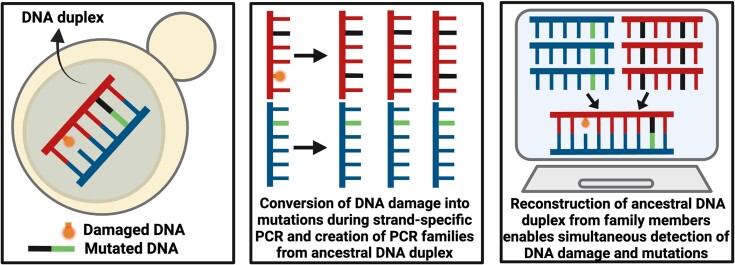
Graphical Abstract
Graphical Abstract.
Introduction
Our genome is exposed to a wide variety of DNA-damaging agents, including radiation, pollution, and metabolic side products (1,2). This damage is a potent source of mutations, which can fuel the evolution of human cancers and initiate the development of genetically inherited diseases (3). Accordingly, researchers and clinicians need sophisticated tools to detect DNA damage and mutations with the utmost sensitivity. These tools could inform strategies aimed at the prevention and treatment of diseases caused by DNA damage and mutation and advance our understanding of the basic biology that underlies these processes.
The most flexible tool to detect DNA-based endpoints is massively parallel sequencing. This technology does not only have the potential to detect DNA damage and mutagenesis, but also quantify them and determine where DNA adducts and mutations are located across the genome. For example, multiple assays are now capable of detecting bulky DNA adducts with single base pair resolution (4–8). However, it has proven difficult to design similar assays for smaller adducts (9). Like bulky adducts, small DNA adducts are important drivers of human aging and disease, and because they are relatively abundant and amenable to translesion synthesis they tend to be highly mutagenic in nature (9). Thus, sensitive assays capable of detecting small DNA adducts are urgently needed. Ideally, these assays would not only detect DNA adducts, but also the mutations they induce. Currently, most researchers split their samples into two different aliquots, one of which is used to detect DNA damage, while the other is used to detect mutations. These aliquots are then processed with different chemicals and enzymes after which each endpoint is detected with a different instrument (10,11). Accordingly, inter-sample variation is inevitable, making it difficult to compare the number of mutations and DNA lesions detected in a truly quantitative fashion.
To address these issues, we developed MADDD-seq, a novel massively parallel sequencing tool that can detect small DNA adducts and mutations simultaneously, using a single assay, a single sample and a single instrument. To do so, MADDD-seq advances on a sensitive double barcoding strategy that was previously used for mutation detection only (12). We then coupled this barcoding strategy to a new bio-informatic pipeline that uses the information encoded by our barcodes to identify mutations and DNA adducts from the same dataset. This pipeline allows mutations to be mapped onto the genome with single base pair resolution, while adducts can be mapped to a single base on a single strand of DNA. We specifically designed MADDD-seq to be a DNA-based sequencing tool, so that it can be applied to any organism of choice. And because it depends on error-prone translesion synthesis during library amplification to detect DNA adducts, we expect that MADDD-seq will be especially useful for the detection of small, highly mutagenic DNA adducts that drive mutagenesis upon mutagen exposure, as well as the endogenous lesions that arise during the natural aging process.
To demonstrate the multi-functional nature of MADDD-seq, we subjected the budding yeast S. cerevisiae to methylnitronitrosoguanidine (MNNG), an alkylating agent known for creating O6-methyl-guanine lesions (O6-meG), a highly mutagenic adduct (13). We then used MADDD-seq to track these adducts and the mutations they induce over a 24-hour time span to determine the kinetics of DNA repair and mutagenesis under various conditions. These experiments allowed us to monitor every step of the mutagenic process with a single assay and gain precise quantitative insight into key parameters that control our risk for mutation and disease.
Materials and methods
MADDD-Seq assay (mutation and DNA damage detection-sequencing)
Concept
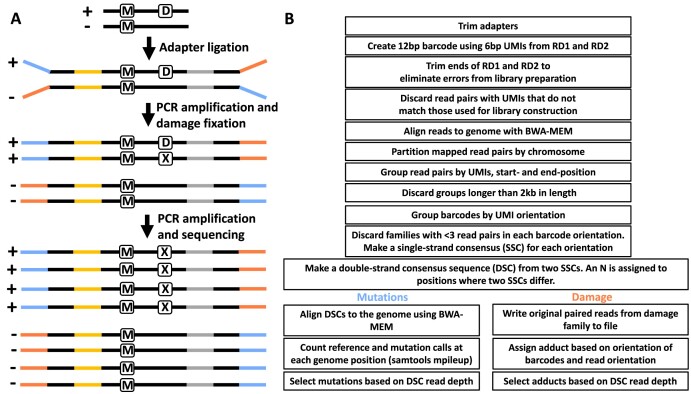
Our custom MADDD-Seq barcoded adapters contain one of 218 known 6-bp unique molecular indices (UMIs) and a forked tail of two non-complementary sequences (blue and orange in Figure 1). A T-overhang on the UMI end of the adapter can be ligated to either end of an a-tailed gDNA fragment. The random pairing of two UMIs ligated to the same fragment (yellow and gray in Figure 1) in combination with the fragment's location on the genome are used to identify each unique DNA fragment. Amplification of this starting fragment generates multiple PCR duplicates that are referred to as ‘family members’ of the ancestral DNA molecule. Mutations present in the ancestral DNA molecule will be present in all PCR duplicates of the top and bottom strand of this duplex. However, errors arising from PCR amplification or next-generation sequencing will only be present in one or a few family members. Therefore, we can distinguish a true mutation, present in the unique starting gDNA fragment, from NGS and PCR-induced errors. Finally, the presence of a DNA lesion in the ancestral DNA duplex will be betrayed by the consistent production of PCR errors that only arise in copies that are made from one of the two ancestral strands.
Figure 1.
Outline of MADDD-Seq method. (A) MADDD-Seq distinguishes mutations from artifacts introduced by sequencing technology or NGS-protocols by confirming that a true mutation (M) is present on both strands of a DNA duplex, while artifacts are only present in one. In addition, MADDD-Seq can identify damaged bases by virtue of the ex vivo mutations that the damage induces during the PCR steps required for library preparation (X, for ex vivo mutation). These mutations arise exclusively and repeatedly on copies that are made from the damaged strand. The + and – symbols refer to the plus and minus strand of a DNA duplex. Directional adapters are depicted as orange and blue lines. UMI’s are depicted as yellow and grey lines (see text for further explanation). (B) Bio-informatic workflow of MADD-seq data analysis
DNA fragmentation
Initial library preparation utilized the NEBNext® Ultra™ II FS Enzyme Mix and NEBNext® Ultra™ II FS Reaction Buffer to fragment, end-repair, and A-tail. However, sonication was implemented to fragment DNA after results suggested enzymatically shearing DNA induced the conversion of single-stranded DNA damage to mutation (Figure 2C, D). In the sonication protocol, 500 ng of genomic DNA in 50 μl was fragmented to ∼300 bp using Covaris S220 (peak power = 175, duty factor = 10.0, cycles/burst = 200, run time = 50 s). Fragment sizes were verified using Agilent TapeStation 4150 using D1000 or D5000-HS reagents.
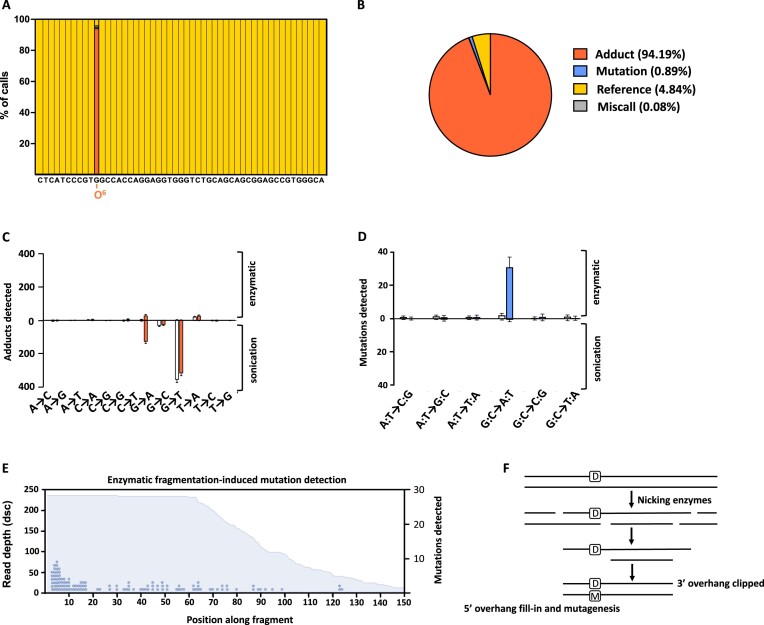
Figure 2.
Detection of an O6-meG lesion in a synthetic oligo. (A) An oligo was synthesized that carries an O6-methyl guanine base at the 11th position and annealed to a complementary strand to create a DNA duplex. This oligo was processed with standard MADDD-seq protocols and pipelines to detect both DNA damage and mutations. MADDD-seq correctly identified an O6-methyl guanine adduct at the 11th position (orange bars) in nearly all sequenced oligos, while it identified undamaged reference bases at all other positions (yellow bars). (B) Deeper analysis demonstrated that at the 11th position, MADDD-seq identified an O6-methyl guanine adduct in 94% of cases, a undamaged reference base in 5% of cases, and a mutation in 0.89% of remaining cases. For (A) and (B), O6-meG adducts are depicted in orange, mutations in blue, reference bases in yellow and miscalls in grey. (C) Isolated DNA was either treated with MNNG (orange bars) or not (white bars), and MADDD-seq was used to detect O6-meG lesions (G→A transitions). This DNA was either sheared by sonication or enzymatic digest. (D) Next, we monitored the same isolated DNA samples for mutations. Treated samples are depicted with blue bars, and untreated samples with white bars. In both (C) and (D), error bars indicate SEM, n = 3. (E) Mutations found in enzymatically fragmented samples are more prevalent towards the start of sequencing reads, suggesting a non-biological assay-induced explanation for the presence of these mutations. The blue surface depicts the read depth of the fragments we sequenced, while the blue dots indicate the number of mutations we detected. One dot indicates one mutation. (F) Schematic of how large overhangs generated by enzymatic fragmentation may convert DNA damage (D) into mutations (M) in pre-PCR gDNA libraries.
Library preparation: end repair and adapter ligation
The fragmented DNA was end-repaired and A-tailed using the NEBNext® Ultra™ II End Prep system. The entire pool of fragmented DNA (∼500 ng) was combined with 7 μl NEBNext Ultra II End Prep Reaction Buffer and 3 μl NEBNext® Ultra™ II End Prep Enzyme Mix and brought to a total reaction volume of 60 μl with TE. The DNA was incubated with the end repair mix at 20°C for 30 min followed immediately by a Beckmann-Coulter AMPureXP bead cleaning at 1.8X to halt enzymatic activity and eluted in 35 ul of 0.1× TE. The NEBNext Ultra II End Prep protocol suggests a 65°C heat inactivation step, which we replaced with this bead cleaning step to minimize heat damage on the libraries.
After the DNA had undergone end repair, we ligated our double-stranded barcoded adapters to the inserts. The 35 μl of end-repaired DNA was combined with 30 μl NEBNext Ultra II Ligation Master Mix, 1 μl NEBNext Ligation Enhancer, and 2.5 μl of 15 μM adapters for a total reaction volume of 68.5 μl. This adapter ligation mix was incubated at 20°C for 15 min before proceeding directly with a Beckmann–Coulter AMPureXP bead cleaning at a concentration of 0.8× to size-exclude surplus adapters and eluting in 20 μl 0.1× TE.
Library quantification
The adapter-ligated libraries were quantified by mass using the Invitrogen Qubit 4 Fluorometer with Qubit 1× dsDNA HS Assay Kit reagents. The fragment size and concentration of each sample was quantified using Agilent's 4150 TapeStation System with D1000 reagents. To get an accurate estimate of the number of adapter-ligated fragments, droplet digital PCR (ddPCR) was performed with primers specific to the u-loop adapters (forward 5′- GACTGGAGTTCAGACGTGTGC-3′ and reverse 5′- CACTCTTTCCCTACACGACGC-3′). The following reagents were combined to form 22.8 μl of template-free master mix for each reaction well: 12 μl QX200 ddPCR EvaGreen® Supermix, 2.4 μl 0.5 μM forward and reverse primers, 8.4 μl nuclease free water (NFW), and 1.2 μl of a 10-fold template dilution series from 1:10 to 1:106. Droplets were generated with Droplet Generation Oil for EvaGreen® using the Bio-Rad QX200 Droplet Generator and amplified with the following cycling parameters: denaturation at 95°C for 5 min followed by 40 cycles of a 30 second denaturation at 98°C and extension at 65°C for 1 minute. This was followed by 5 min at 4°C then a final step of 5 min at 90°C before cooling again at 4°C. Droplets were analyzed by the Bio-Rad QX100 Droplet Reader.
Amplification for Illumina sequencing
Based on quantification by ddPCR, 3 million molecules were amplified using indexed primers for multiplexing samples during sequencing with an adaptation of NEBNext's Multiplex Oligos for Illumina universal forward primer (5′-AAT GAT ACG GCG ACC ACC GAG ATC TAC ACT CTT TCC CTA CAC GAC GC-3′) and indexed reverse primers, 5′-CAA GCA GAA GAC GGC ATA CGA GAT XXX XXX GTG ACT GGA GTT CAG ACG TGT GC-3′), where the bases ‘XXX XXX’ correspond to indices 1,2,4,5,6,7,12,15,16, and 19. Index primers were selected from a combination of Index Primers Set 1 and Set 2 based on the NEB index pooling guide. The primers used are identical to NEBNext's Multiplex Oligos for Illumina except for the removal of 10 bp from the 3′ end of each primer complementary to the double-stranded portion of the adapters to prevent these primers from annealing with molecules from the opposite strand and conflating the sequence-based strand distinction in our assay.
The amplification reaction contained 25 μl NEBNext Q5 2× MM, 2.5 μl 10 μM NEBNext Universal Primer, 2.5 μl 10 μM NEBNext Index Primer, an aliquot of 3 million copies of template, and NFW to bring the total volume to 50 μl. Cycling parameters for amplification were as follows: initial denaturation at 98°C for 30 s, followed by 15 cycles of denaturation at 98°C for 10 s and extension at 65°C for 1 min. The appropriate extension time and temperature was determined with the help of oligos that do, or do not contain an O6-meG lesion at the 11th position. By amplifying damaged and undamaged oligos side by side for 15 cycles, and then measuring their relative abundance through sequencing, we were able to determine that both the damaged and undamaged DNA molecules amplify with equal efficiency under these conditions. A final step of 65°C for 5 min was performed prior to sample cooling at 4°C. Two successive AMPure XP bead purification of PCR products were performed at 0.7× concentration, and the samples were eluted in a final volume of 20 μl 0.1× TE.
Amplified library quantification and re-amplification
Amplified libraries were quantified by Invitrogen's Qubit 4 Fluorometer with Qubit 1× dsDNA HS Assay Kit reagents and Agilent's 4150 TapeStation System with D5000 High Sensitivity reagents. ddPCR was performed using primers complementary to NEBNext's Multiplex Oligos for Illumina: forward 5′-AAT GAT ACG GCG ACC ACC GA-3′ reverse 5′-CAA GCA GAA GAC GGC ATA CGA-3′ and a FAM fluorescent probe complementary to the Illumina primers to specifically measure the number of DNA fragments that could be sequenced (5′-/56-FAM/CCC TAC ACG /ZEN/ACG CTC TTC CGA TCT/3IABkFQ/-3′). The reaction mixture contained 12 μl ddPCR Master Mix for Probes, 3 μl 2 μM DPL probe, 2.16 μl 10 μM primers, 1.2 μl template along a 1:10 – 1:106 dilution gradient, and NFW to 24 μl. After droplet generation using the QX200 droplet generator and ddPCR Droplet Generation Oil for Probes (Bio Rad), reactions were heated at 95°C for 10 min, then denatured at 95°C for 30 s and extended at 65°C for 1 minute for 40 cycles before heating at 98°C for 10 min and cooling at 4°C. Droplets were analyzed on the Bio Rad QX100 Droplet Reader.
Based on ddPCR quantification, samples that were not concentrated enough to sequence were re-amplified to target ideal sequencing concentration. The remaining sample volume was amplified in a 50 μl reaction mixture along with 2 μl NEBNext Q5 2X MM, 5 μl 10 μM library primers (forward 5′-AAT GAT ACG GCG ACC ACC GA-3′ reverse 5′-CAA GCA GAA GAC GGC ATA CGA-3′), and NFW to 50 μl. Cycling parameters for amplification were as follows: initial denaturation at 98°C for 30 s, followed by a variable number of amplification cycles of denaturation at 98°C for 10 s and extension at 65°C for 1 minute targeting a final concentration of 1.5 nM. A final step of 65°C for 5 min was performed prior to sample cooling at 4°C. Libraries were purified using 0.9× AMPure XP bead and eluted in 15 μl 0.1× TE. Library quantification was reassessed using TapeStation, Qubit, and ddPCR for Probes as described directly above. Re-amplification and quantification were repeated as necessary until libraries reached approximately ∼0.2–1.0 nM. Target concentrations are optimized to be the lowest possible given the total quantity of DNA in order to sequence as much of the library as possible. Samples were then multiplexed and run on a MiSeq Nano Kit using custom primers specific to the NEBNext Index primers (Read 1 Primer: 5′-ACA CTC TTT CCC TAC ACG ACG CTC TTC CGA TCT-3′, Index Primer: 5′-GAT CGG AAG AGC ACA CGT CTG AAC TCC AGT CAC - GCC AAT - ATC TCG TAT GCC GTC TTC TGC TTG-3′, Read 2 Primer: 5′-GTG ACT GGA GTT CAG ACG TGT GCT CTT CCG ATC T-3′). Based on MiSeq clustering and read distribution, sample volumes were adjusted for equal distribution of reads, pooled, and sequenced on a NextSeq 2000.
UDSBC adapter preparation
U-loop double stranded barcode adapters (5′-TGA CTA GAT CGG AAG AGC ACA CGT CTG AAC TCC AGT C dU A CAC TCT TTC CCT ACA CGA CGC TCT TCC GAT CTA GTC A-s-T-3′) were annealed for 5 min at 95°C then cooled quickly on ice. To prevent cleavage at sites of damage on gDNA, 24 μl of 15μM annealed u-loop adapters were digested prior to ligation using 2 μl USER enzyme in a 40 μl reaction with 1× CutSmart Buffer for 15 min at 37°C. Digested adapters were bead cleaned using AMPure XP beads at 1.8× concentration and eluted in 16 μl 0.1× TE. Note that pre-digestion of adapters results in a reduction in ligation efficiency and that future library preparations will exclude this step.
Bio-informatic processing
Raw data was processed using standard protocols for quality control. Briefly, after trimming of adapters, the barcodes were identified and trimmed to eliminate artifacts that may arise from library preparation. Sequences were then aligned to the genome with BWA-MEM and grouped by the orientation of their UMI. Families were only included for further analysis if they contained 3 or more read pairs in each barcode orientation (corresponding to copies of the top and bottom strand of a DNA duplex). The consensus sequences of these read pairs were then processed through either a mutation or DNA damage detection pipeline. The data derived from these pipelines was then parsed again to exclude sequences corresponding to rDNA and transposable elements. The multicopy nature of these sequences confounds mutational analyses because individual copies of these genes may carry pre-existing mutations. For similar reasons, we excluded mutations detected at more than 1 time point, as they must have existed before our 24-hour experiment started.
Yeast culture
Single colonies were inoculated in YAPD and incubated overnight at 30°C in a rotating wheel. In the morning, the optical density (OD600nm) of each culture was measured using Thermo Scientific's Nanodrop 2000C and cells were re-inoculated at an OD600 0.05–0.1 in 50 ml YAPD flasks and incubated in an orbital shaker at 30°C. Cells were then grown to an OD of 0.25–0.5 and either harvested or arrested with 50 μg/ml α-mating factor. After 2.5 h, the cells were visualized under a microscope to confirm they were arrested, and then treated (or not) for 40 min with 10 μg/ml MNNG. After treatment, the cells were washed 3 times in PBS with α-mating factor to remove mutagenic compounds and re-inoculated in YAPD with α-mating factor to maintain arrest over the recovery period.
Concept
Massively parallel sequencing tools have revolutionized modern medicine (14). They exposed the genetic heterogeneity of human cancers, identified mutations responsible for inherited diseases, and opened the door for personalized medicine (15). In addition, these tools can be used to detect mutations that are only present in one or a small number of cells (10). Although rare, these mutations provide valuable insight into the origins of human cancers (16), the basic biology that underpins human aging (17), and the impact of environmental mutagens on the human genome (18). To detect random mutations, massively parallel sequencing assays had to be modified to filter out PCR artifacts that could otherwise be interpreted as mutations (19,20). One of the most important contributors to these artifacts is DNA damage, which increases the error rate of DNA polymerases during the PCR amplification steps that are inherent to library preparation. To filter out these artifacts, multiple laboratories designed a double-stranded barcoding strategy that tags both strands of a DNA duplex with a specific barcode (21). These barcodes allow a DNA duplex to be reconstructed from its individual DNA strands from raw sequencing data. The rationale behind this strategy is that if a mutation is present in a DNA fragment, it should be present in both strands of the DNA duplex, while an artifact (such as ex vivo mutations introduced by DNA damage during PCR amplification) are present in only one (21).
The concept behind MADDD-seq is to exploit the ability of DNA damage to introduce ex vivo mutations into massively parallel sequencing libraries for detection purposes. Because ex vivo mutations are only introduced in PCR copies made of the damaged strand while copies of the undamaged strand remain artifact-free, it should be possible to detect DNA adducts if PCR copies from the top and bottom strand of a DNA duplex can be distinguished from each other. To do so, we designed a modified set of double-stranded barcodes (Figure 1A) that we coupled to a custom bio-informatic pipeline that keeps track of ex vivo mutations that arise in copies from each strand in a DNA duplex (Figure 1B). Importantly, this strategy leaves the mutation detection component of ‘duplex sequencing’ intact, so that MADDD-seq can detect both mutations and DNA damage simultaneously, with a single assay on a single sample.
Our barcoding strategy resembles the forked adapters used in traditional duplex sequencing techniques. However, MADDD-seq adapters carry different sequences on their forked ends, and alternate in their orientation towards the UMIs present on each molecule. For example, in the schematic presented in Figure 1, the orange adapter is always adjacent to the purple UMI on the top strand but paired with the green UMI on the bottom strand. Thus, molecules derived from the top strand always read blue fork – yellow UMI – insert – grey UMI – orange fork, while molecules from the bottom strand read blue fork – grey UMI – insert – yellow UMI – orange fork. Our bio-informatic pipeline uses this information to distinguish between copies that are made from the top or bottom strand of a DNA duplex, so that DNA damage can be localized to a single base, in a single strand of the original DNA duplex.
Results
To test our barcoding strategy, we synthesized a 50 bp double-stranded oligonucleotide that carries an O6-meG adduct at the 11th position. O6-meG lesions are small but highly mutagenic adducts that arise due to endogenous and exogenous processes alike (22) and are associated with a wide variety of spontaneous and environmental cancers (22,23). In addition, they were recently implicated in the etiology of Alzheimer's disease (24). Thus, a sensitive assay for the detection of O6-meG adducts is desirable and could be an important factor for disease prognosis, tumor grading, and the prediction of treatment efficacy. We found that MADDD-seq correctly identified an O6-meG adduct at the 11th position in 94% of the sequenced oligonucleotides (Figure 2A, B). At that position, our bio-informatic pipeline consistently identified G →A mutations in copies that were made of the damaged strand, the primary mutation induced by O6-meG. In contrast, copies of the undamaged strand remained error-free. In 5% of cases, an undamaged base was called, while the damaged base was mistaken for a mutation in 0.89% of cases. It is possible that the relative inaccuracy of inserting an O6-meG base at this position during oligo synthesis accounts for some of the undamaged base calls, as the manufacturer only guarantees that >85% of oligos will contain the desired O6-meG adduct. Regardless, this data demonstrates that MADDD-seq allows O6-meG lesions to be detected with at least 94% accuracy.
After acute exposure to a mutagenic compound, O6-meG adducts are randomly distributed across a complex genome that contains countless structural features. To determine whether MADDD-seq can identify O6-meG lesions under those conditions as well, we treated isolated DNA from arrested yeast cells with MNNG, an alkylating agent that creates O6-meG lesions. We used isolated DNA for this experiment to prevent MNNG from inducing mutations, a strategy that has the added benefit of allowing us to test whether MADDD-seq can correctly distinguish between O6-meG lesions and mutations. Consistent with this idea, we found that after exposure, MADDD-seq recorded a substantial increase in O6-meG lesions, while no increase was detected in mutagenesis. Moreover, MADDD-seq detected O6-meG adducts on all 16 chromosomes, indicating that it can be used to detect O6-meG adducts in a true genome-wide fashion. We did discover though, that MADDD-seq was only capable of distinguishing between O6-meG adducts and mutations if the DNA was sheared by sonication (Figure 2C, D). If the DNA was fragmented enzymatically, MADDD-seq only reported a small increase in O6-meG adducts and a large increase in mutations (Figure 2E). These mutations were biased toward the end of sequenced DNA fragments, suggesting a non-biological explanation. It is possible that these artifacts were created by single-stranded overhangs and gaps generated by the enzymatic fragmentation mixture, which were then filled in by the DNA polymerases that are also included in the mixture, thereby creating mutations opposite O6-meG bases (Figure 2F). Thus, the DNA fragmentation method is essential to the success of O6-meG detection by MADDD-seq. Surprisingly though, MADDD-seq also detected a second DNA damage signature in the samples that were sheared by sonication characterized by G→T mutations. This signature was present in both damaged and undamaged DNA samples, indicating that it was not related to MNNG treatment. Instead, it must have been present in the DNA prior to fragmentation, or the result of sonication itself.
After establishing the optimal protocol to detect O6-meG adducts with MADDD-seq, we wanted to test its ability to detect DNA damage and mutations simultaneously. To do so, we mimicked the acute exposure of an organism to a mutagen by treating rapidly dividing yeast cells with MNNG for 40 min, washed the cells, and let them recover for 24 h while we tracked the presence of DNA adducts and mutations in the cells (Figure 3A). As expected, we found that MADDD-seq identified a large increase in O6-meG adducts immediately after exposure (t = 0) by their distinct G→A damage signature. These adducts covered the entire genome in a relatively unbiased fashion, affecting protein-coding regions, intragenic sequences, introns, untranslated regions, and regulatory elements alike (Figure 3B–D). A full list of all the damaged bases detected in this study is provided in Supplementary Table S1. During the 24-hour recovery period, these lesions rapidly declined and returned to baseline levels once the experiment was concluded. Most likely this rapid decline is the result of the dual activity of DNA repair and DNA replication. Interestingly, we did not detect the additional G→T damage signature observed in naked DNA, suggesting that this signature is not the result of sonication. However, we did observe a relatively weak DNA damage signature characterized by C→T mutations. At first glance, it was unclear whether this signature was directly related to MNNG exposure, because it did not appear until 2 h after MNNG exposure had ended (t = 2) and was lost after the next time point (t = 6). Because MADDD-seq reports a base as damaged when mutations accumulate in copies made from only one strand of a DNA duplex, we hypothesized that the C →T damage signature could be the result of thymine misincorporation events opposite O6-meG lesions during the first round of DNA replication after MNNG exposure. If so, these misincorporations would lead to an O6-meG:T mismatch that would be converted into a regular G:T mismatch after DNA repair. This misincorporated thymine would then be reported as a damaged base because the resulting C→T mutation would only be present in copies made from one of the two strands of a DNA duplex. A second round of DNA replication would then result in the loss of the C to T damage signature, as both strands of the DNA duplex would now contain a mutant base.
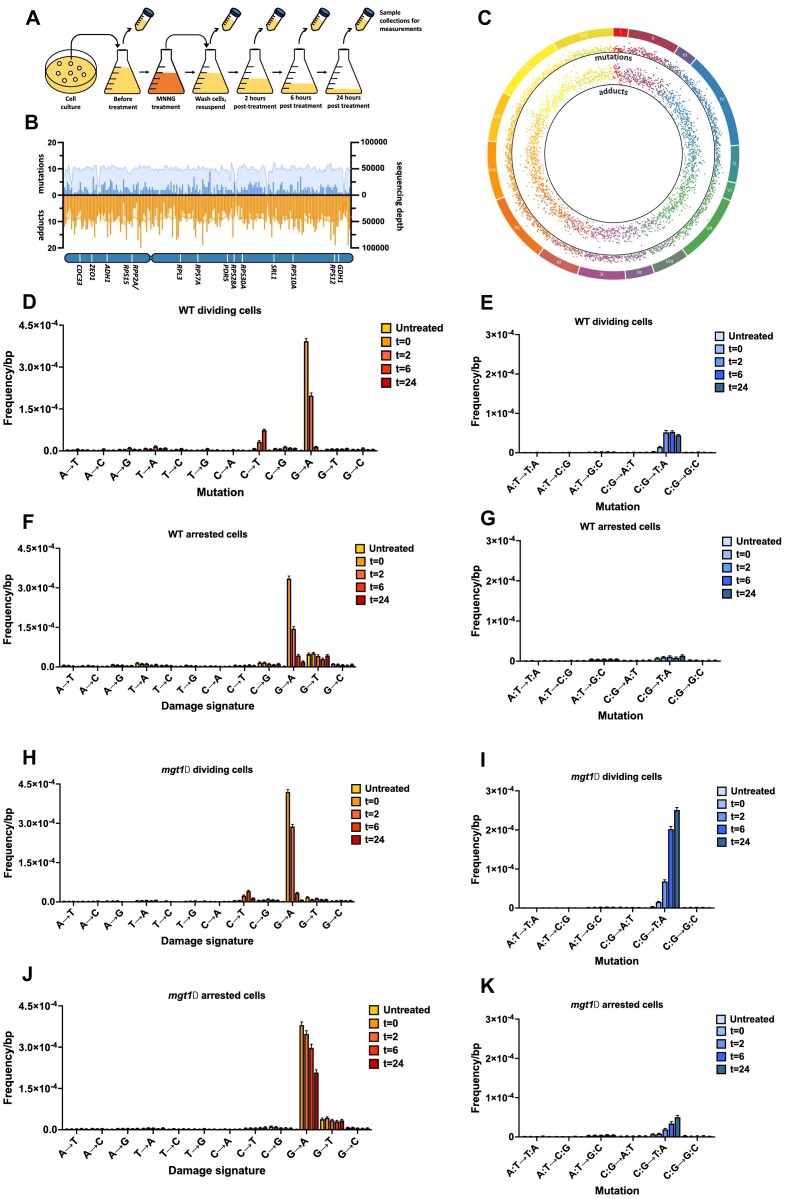
Figure 3.
Simultaneous detection of DNA damage and mutations in yeast cells with MADDD-seq. (A) Schematic representation of experimental procedures. Single colonies were inoculated in liquid medium and expanded into a 250ml culture. Over the course of 24 h, multiple 50ml aliquots of cells (conical tubes) were collected at different time points to detect DNA damage and mutagenesis by MADDD-seq. (B) Circos plot of all mutations and adducts detected in this study. The outer ring depicts the chromosome and the location along the chromosome that mutations and adducts were detected at. Each dot in the inner ring depicts the number of adducts detected in 5000 bp intervals. The further away from the center of the plot, the more adducts were detected. The middle ring depicts the number of mutations detected in the same 5000 bp intervals. The further away from the center of the plot, the more mutations were detected. (C) MADDD-seq coverage and endpoint detection across chromosome XV. Coverage is depicted by the colored surface, while mutations are depicted as orange bars, and adducts as and blue bars, binned in 5,000bp intervals. This plot contains all the mutations and adducts detected in this study. (D, F, H, J) Frequency of DNA damage signature in the nuclear genome of dividing and arrested cells, with or without deletion of MGT1. (E, G, I, K) Frequency of mutations in the nuclear genome of dividing and arrested cells, with or without deletion of MGT1. Error bars indicate upper and lower limits of 95% confidence intervals for the fraction, n = 1.
To provide evidence for this hypothesis, we analyzed our dataset with the second arm of our bioinformatic pipeline, which is designed to detect mutations. A full list of all the mutated bases detected in this study is provided in Supplementary Table S2. Consistent with the mutagenic nature of O6-meG adducts we found that the mutation frequency of the treated cells rapidly increased over the first 2 h after treatment. Moreover, this increase was completely driven by CG:TA mutations (Figure 3E), in accordance with the mutation spectrum of O6-meG adducts and the G→A and C→T damage signatures we observed. If the C→T damage signature is indeed the result of misincorporation events during DNA replication, we reasoned that this signature should not be present in cells that are in a non-dividing state. Therefore, we arrested the cells with α-mating factor, treated them with MNNG and tracked the presence of mutations and DNA damage over a 24-h time course. Consistent with our hypothesis, we found that the arrested cells accumulated similar numbers of O6-meG lesions compared to dividing cells (t = 0, Figure 3F), but the C→T damage signature did not appear. In fact, relatively few mutations arose over the entire 24-hour recovery period (Figure 3G), underscoring the idea that cell division and misincorporation events during DNA replication are essential steps for the conversion of O6-meG lesions into mutations. Similar observations were previously made for other DNA lesions (25), indicating that non-dividing cells are protected from mutagenesis after mutagen exposure. We did observe the G→T damage signature previously observed on naked DNA though. Since this signature is only observed in DNA from arrested cells, it could have a biological origin that is related to cell cycle arrest by α-mating factor. Interestingly, a whole transcriptome analysis of arrested and dividing yeast cells indicates that arrested cells display a 25-fold increase in YAP1 expression, a critical transcription factor that controls the response of yeast cells to oxidative damage (26) (Table 1). Accordingly, numerous genes that are controlled by YAP1 and counteract oxidative stress are upregulated in arrested cells, including the glutathione synthetases GSH1 (4.4-fold) and GSH2 (7.8-fold), the glutathione transferases GTT1 (2.2-fold) and GTT2 (7-fold), the glutathione peroxidases GPX1 (2.5-fold) and GPX2 (12.5-fold), the anti-oxidant enzymes SOD1, SOD2 and CTT1, and the DNA repair protein OGG1 (2.5-fold). Thus, one possibility is that this damage signature is the result of oxidative stress. Notably, 8-oxo-guanine lesions mispair with adenine during DNA replication (27), which could result in a G→T damage signature seen in arrested cells.
Table 1.
Expression of genes associated with oxidative stress
| Gene | Function | Change in arrested cells |
|---|---|---|
| SOD1 | Cytosolic superoxide dismutase | +23.5-fold |
| YAP1 | Transcription factor | +19.9-fold |
| GPX2 | Glutathione peroxidase | +12.5-fold |
| SOD2 | Mitochondriall superoxide dismutase | +8.7-fold |
| GSH2 | Glutathione synthetase | +7.8 fold |
| GTT2 | Glutathione S-transferase | +7.0-fold |
| DUG1 | Glutathione degradation | +6.6-fold |
| GSH1 | Glutathione synthetase | +4.7-fold |
| GCG1 | Gamma-glutamyl cyclotransferase | +4.1-fold |
| CTT1 | Catalase | +3.6-fold |
| OGG1 | 8-Oxo-guanine glycosylase/lyase | +2.5-fold |
| DUG2 | Component of glutamine amidotransferase | +2.4 fold |
| GTT1 | Glutathione S-transferase | +2.2-fold |
| GPX1 | Glutathione peroxidase | +1.5-dold |
| DUG3 | Component of glutamine amidotransferase | –5.4-fold |
| GLR1 | Glutathione reductase | –20.0-fold |
A whole transcriptome analysis indicated that genes associated with oxidative stress are upregulated in arrested cells compared to rapidly dividing cells. These genes included the key transcription factor YAP1, the antioxidant enzymes SOD1, SOD2 and CTT1, various lynchpins of glutathione metabolism, and the DNA repair protein OGG1.
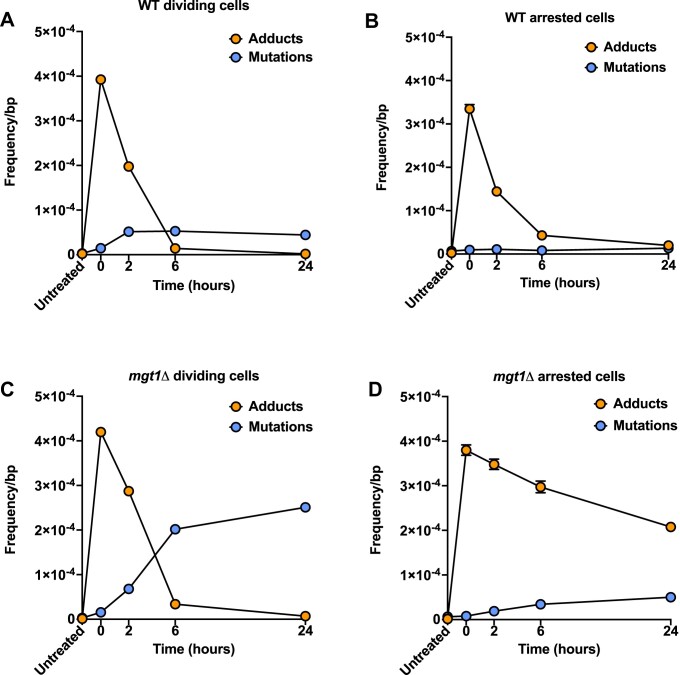
Next, we wanted to test the ability of MADDD-seq to detect DNA damage and mutations in a medically relevant scenario. To do so, we mimicked hypermethylation of the MGMT promoter in brain tumors by deleting MGT1 in dividing yeast cells and treated the cells with MNNG. After exposure, the mgt1Δ cells accumulated a similar amount of O6-meG lesions compared to WT cells, indicating that the presence or absence of Mgt1p has little effect on the total number of lesions that arise in yeast cells (Figure 3H). However, Mgt1Δ cells converted these lesions into mutations at a higher rate compared to WT cells (Figure 3I), underscoring how important Mgt1p is for the prevention of mutagenesis by O6-meG lesions. These measurements illustrate how MADDD-seq could be deployed in biomedical research or the clinic to investigate the consequences of genetic and epigenetic alterations to MGMT in human cells. As expected, the mgt1Δ cells also displayed the C→T damage signature that is indicative of G:T mismatches. Surprisingly though, this signature was less pronounced compared to WT cells. Most likely, this reduction is caused by deletion of the Mgt1 protein, which prevents O6-meG:T mismatches from being converted into G:T mismatches. As a result, both strands will generate mutant copies during PCR amplification, causing O6-meG:T mismatches to be recorded as mutations. Accordingly, MADDD-seq may slightly overestimate the mutation frequency during the first time points after MNNG treatment in mgt1Δ cells.
In addition to dividing cells, like the cells from a brain tumor, non-dividing cells can be deficient for MGMT as well. For example, it was recently shown that female patients with non-familial cases of Alzheimer's disease display hypermethylation of the MGMT promoter, leading to reduced MGMT expression, while males do not (24). Potentially, this reduced expression could help explain why females are at greater risk for Alzheimer's disease compared to males. To mimic MGMT hypermethylation in neurons and explore the impact of reduced MGMT expression on non-dividing cells, we arrested mgt1Δ cells with α-mating factor and treated them with MNNG. Consistent with the critical role that Mgt1p plays in the repair of O6-meG adducts, we found that these cells retained O6-meG lesions for an extended period of time (Figure 3J). However, similar to WT cells, these lesions induced less mutations compared to dividing cells (Figure 3K). Together, these observations underscore the importance of Mgt1p for the repair of O6-meG lesions in non-dividing cells and the relative resistance of arrested cells to DNA damage induced mutagenesis.
One exciting aspect of MADDD-seq is that it can detect mutations and O6-meG lesions simultaneously, allowing for direct comparisons between these endpoints. To do so, we generated additional plots that directly compare the frequency of the O6-meG lesions created by MNNG to the CG:TA mutations they induce (Figure 4A–D). All other lesions and mutations were excluded from analysis. These plots illustrate that in dividing WT cells O6-meG adducts were present at a frequency of 3.92 × 10−4/bp after exposure (t = 0), which increased the mutation frequency 16-fold to 4.42 × 10−5/bp over a 24-hour time span (t = 24, Figure 4A). This comparison indicates that 11% of the O6-meG lesions present at t = 0 were converted into mutations, while 89% were repaired. The efficiency with which mutations were converted into mutations was substantially higher in mgt1Δ cells. Even though these cells carried a nearly identical number of lesions immediately after exposure (4.2 × 10−4/bp), they converted 60% of these lesions into mutations, raising the mutation frequency 85-fold to 2.5 × 10−4/bp (Figure 4B). These straightforward calculations demonstrate how simultaneous detection of DNA damage and mutations allows mutagenic endpoints to be quantified in a highly direct, informative manner. Our dataset was equally informative when we examined arrested cells. In arrested WT cells, a substantial number of O6-meG lesions were created after MNNG exposure as well (3.3 × 10−4), but in contrast to dividing cells, the mutation frequency did not even double over the ensuing 24-hour time period (Figure 4C). We estimate that only 4% of lesions were converted into mutations, further highlighting that arrested cells are protected from mutagenesis by O6-meG lesions. In this context, it should be noted that approximately ∼6% of cells escaped arrest after 24 h of α-mating factor exposure and re-entered the cell cycle (Supplementary Figure S1). Accordingly, some of these mutations may have occurred in cells that re-entered the cell cycle. We observed similar protection from mutagenesis in arrested mgt1Δ cells (Figure 4D), although these cells accumulated more mutations than WT cells, consistent with their inability to repair O6-meG lesions in an efficient manner (28).
Figure 4.
Direct comparisons between DNA damage and mutagenesis. (A–D) Frequency of O6-meG lesions and CG:TA mutations measured over a 24-h time span after MNNG treatment in WT cells and Mgt1Δ cells that are either in a dividing or arrested state. Error bars indicate upper and lower limits of 95% confidence intervals of the fraction, n = 1. Note that for most data points these error bars are too small to be depicted.
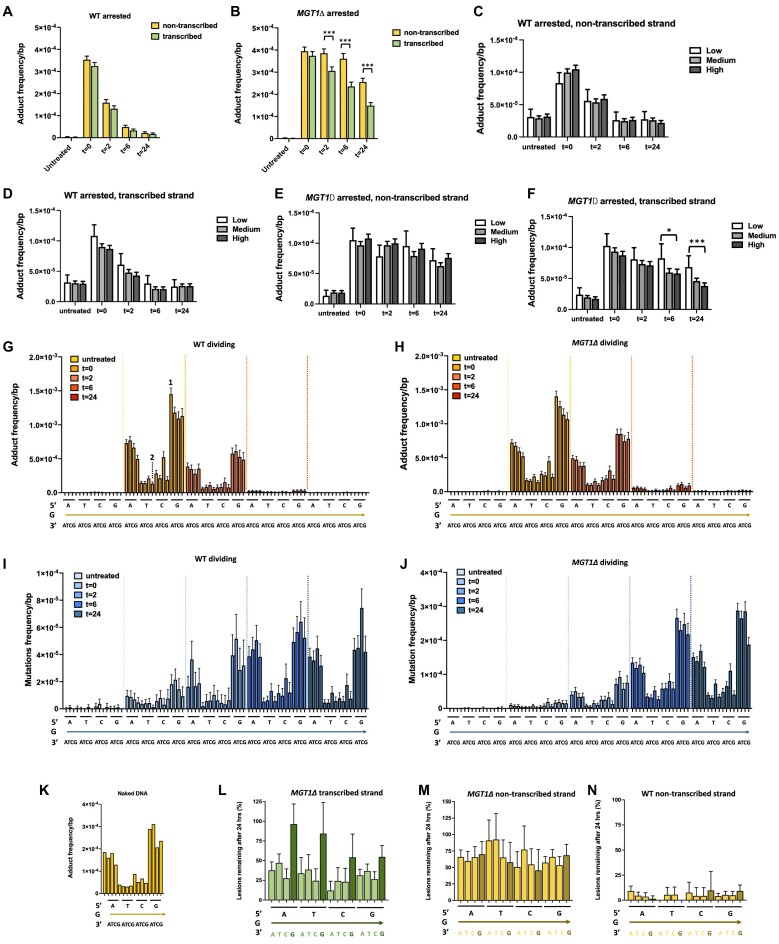
The experiments described above demonstrate that MADDD-seq can identify mutations and DNA damage across the genome in a massively parallel fashion. As a result, it may be possible to mine MADDD-seq datasets for known and unknown variables that control the efficiency with which the genome is either repaired or mutated. One open-ended observation we made was that arrested cells that lack Mgt1p still display evidence of DNA repair, suggesting that a second DNA repair pathway may compensate for Mgt1p deletion. To identify this pathway, we first dividied the genome into transcribed and untranscribed regions and then compared the rate with which O6-meG lesions are repaired between regions. Interestingly, we found that in mgt1Δ cells, repair of O6-meG lesions is more pronounced on the transcribed strand of protein-coding genes compared to the non-transcribed strand, a distinction that is not visible in WT cells (Figure 5A, B). This observation suggests that in the absence of Mgt1p, transcription-coupled DNA repair plays an important role in removing O6-meG lesions from the genome. To explore this possibility further, we analyzed the transcriptome of treated and untreated yeast cells by RNA-seq and found that genes with higher expression levels indeed display faster DNA repair compared to genes that are rarely transcribed, specifically on the transcribed strand. Again, no such distinction was visible in WT cells, in which most O6-meG lesions are repaired is performed by Mgt1p (Figure 5C–F).
Figure 5.
Adduct repair and mutations induction as a function of genetic context and transcription level. (A, B) O6-meG lesions are removed faster from the transcribed strand compared to the non-transcribed strand in arrested Mgt1Δ cells, but not in WT cells. (C–F) Based on transcriptome analysis (n = 6 for all conditions), genes were divided into 3 bins: low, medium and high transcription level and the frequency of O6-meG lesions was monitored across these three bins over a 24-h timespan. We found that O6-meG lesions were removed faster from highly transcribed genes compared to rarely transcribed genes, but only in the transcribed strand of arrested Mgt1Δ cells. (G–J) The frequency of mutations (blue bars) and O6-meG lesions (orange bars) and were monitored over a 24-h timespan as a function of the genetic context in which the damaged base was present. The base that flanks the O6-meG lesion on the 5′ side is depicted immediately underneath the graph. The damaged guanine base itself is depicted underneath the 5′ base (and is present in all triplets, as depicted by the arrow), while the base flanking the O6-meG lesion on the 3′ side is depicted below the arrow. For example, at t = 0, O6-meG lesions are most common at guanine bases that are flanked on its 5′ side by another guanine, and on its 3′ side by adenine (labeled by the number 1). In contrast, guanine bases that are flanked on its 5′ side by thymine and on its 3′ side by guanine are the least likely to carry an O6-meG lesion (labeled by the number 2). For comparisons between adducts and lesions, please compare the adducts at t = 0 (when the maximum number of lesions are present) to the mutations at t = 24 (when all these lesions have been fixed into mutations). (K) Naked DNA treated with MNNG displays similar patterns of O6-meG accumulation compared to cells (H and J). The transcription-coupled DNA repair process that removes O6-meG lesions from the genome prefers a genetic context in which a guanine base (dark green) is not present on the 3′ side of the damaged base. This preference is not observed in the non-transcribed strand of Mgt1Δ cells (L), the transcribed strands of WT cells (M), or the transcribed strand of WT cells (N). For (L), (M) and (N) the 3′ guanine base is labeled in a darker color. Statistical tests for significance were performed using tests of equal proportion with Yates continuity correction. * P< 0.05, ** P< 0.01, *** P< 0.001. For all conditions, n = 1. Error bars represent 95% confidence interval of the fraction.
In addition, we analyzed our dataset to identify new variables that link DNA damage to mutations. Mutations are frequently found to arise in a non-random fashion after mutagen exposure, with the rate of mutation changing as a result of genetic context (29). We found that our data displays a similar trend, with mutations arising more frequently on O6-meG lesions that are flanked by a purine on their 5′ side compared to a pyrimidine (Figure 5G, H). However, it is unknown what mechanism is responsible for this asymmetric distribution. To identify this mechanism, we examined the initial distribution of O6-meG lesions across the genome after MNNG exposure and found that O6-meG lesions preferentially arise on guanine bases flanked on their 5′ side purines (Figure 5I, J). We observed a similar pattern on naked DNA treated with MNNG, suggesting that this pattern is directly related to the primary sequence of the genome (Figure 5K). Together, these observations suggest that the genetic context in which mutations arise is not dictated by a preference of Mgt1p for the repair of O6-meG lesions in a specific genetic context, or preferential misincorporation by replicative polymerases, but rather the specificity with which O6-meG lesions accumulate on the genome. We did observe though, that in contrast to Mgt1p, the transcription-coupled DNA repair process that compensates for MGT1 deletion does display a subtle preference for certain genetic contexts. Most notably, we found that O6-meG lesions that are flanked on their 3′ side by a second guanine are repaired less efficiently compared to O6-meG lesions flanked by adenine, cytosine or thymine (Figure 5L). Potentially, this preference could skew the mutation profile of cells that lack Mgt1p. Consistent with the idea that this preference is specific to transcription-coupled DNA repair, we did not observe this discrepancy on the non-transcribed strand of mgt1Δ cells (Figure 5M) or in the transcribed strand of WT cells (Figure 5N). Taken together, these observations demonstrate how MADDD-seq datasets can be used to make detailed observations that explore key aspects of the relationship between DNA damage, DNA repair and mutagenesis.
Discussion
Mutations play an important role in human aging and disease. However, because they are rare and randomly distributed across the genome, they are exceedingly difficult to detect. To solve this problem, a number of highly sophisticated genome-wide detection techniques have been developed that greatly improve the detection of mutations, including Cypher-seq (12), circle-sequencing (30), o2n-sequencing (31), SMM-seq (32), RADAR-seq (33) and duplex sequencing (19). However, to fully understand the role of mutagenesis in human pathology it is equally important to design tools that can detect DNA damage, the primary mechanism by which mutations arise. Like mutations, DNA adducts are rare and randomly scattered across the genome, but on top of that, DNA damage also comes in many shapes and sizes, with each lesion representing a new challenge to overcome (11). Despite these challenges, multiple highly advanced and often complementary techniques have been developed over the past decades. For example, mass spectrometry (34) and radioactive labeling techniques (35) now allow researchers to determine the total amount of DNA damage present in a biological sample, while the precise locations of lesions can be identified with lesion-specific sequencing techniques using antibodies and modified enzymes (4,6–8,36–47) or long-range PCR assays (48–51). It is important to note that the sensitivity of these techniques is highest when they target bulky lesions, while smaller lesions tend to be missed. For example, antibodies raised against DNA adducts tend to work best when they target bulky lesions that are easy to discriminate from undamaged bases, while long-range PCR reactions require lesions to block DNA polymerases during DNA synthesis, which is more likely to happen in the case of bulky adducts (52). In contrast, small lesions are harder to discriminate from undamaged bases and allow for more efficient translesion synthesis during DNA synthesis, a feature that enhances their mutagenicity. To address this issue, we developed MADDD-seq, an assay that is specifically designed to detect small lesions that are highly mutagenic and allow for efficient translesion synthesis. In addition, MADDD-seq is capable of simultaneous detection of the mutations that are induced by these lesions.
In the proof-of-principle experiments, we describe here, we demonstrate that MADDD-seq can identify O6-meG lesions and mutations in a true genome-wide fashion. Importantly, O6-meG lesions are associated with a wide variety of human cancers. For example, the promoter of the gene that encodes the DNA repair protein MGMT is hypermethylated in 50% of grade IV glioblastomas, 85% of thyroid cancers and 70% of colorectal cancers (53,54), suggesting that O6-meG lesions are the primary source of the mutations that drive these cancers. In addition, MGMT is hypermethylated in female (but not male) patients with Alzheimer's disease. which could contribute to the increased mutagenesis and apoptosis seen in the hippocampal neurons of patients (55), as well as the sexual dimorphism of the disease itself. Thus, a sensitive assay for the detection of O6-meG lesions and the mutations they induce could be an important factor for disease prognosis, tumor grading, and the prediction of treatment efficacy. Here, we demonstrate that MADDD-seq identifies clear differences between the number of DNA lesions and mutations present in WT and mgt1Δ cells, outlining the parameters that could inform aspects of clinical care.
Because MADDD-seq creates large data sets that allow for direct comparisons between DNA damage and mutagenesis, MADDD-seq can also be a valuable tool for basic scientists. For example, we show here that MADDD-seq data can be parsed to examine the kinetics of DNA repair across the genome, as well as the rate at which lesions are fixed into mutations. Further filtering of this data allowed us to determine the functional and sequence-specific parameters that control these processes. For example, after treating WT and mgt1Δ cells yeast cells with MNNG, we were able to reveal the distribution of O6-meG lesions across the genome, the kinetics with which they were repaired and the percentage of lesions that were converted into mutations. With the help of a G→A damage signature that identifies O6-meG lesions, a C→T damage signature that identifies the first mutant base misincorporated opposite an O6-meG lesion, and CG:TA mutations that represent the final mutated DNA duplex, we were able to capture every step related to O6-meG-mediated mutagenesis. Although these observations clearly demonstrate the potential of MADDD-seq technology to make precise biological observations, it should be noted that this study was primarily designed to validate our assay and to provide a snapshot of what MADDD-seq can do. For this reason, all of our biological samples were processed with an n of 1, which means that the biological findings we report here are preliminary in nature and further analysis is required to confirm them.
These observations hint at various basic biological processes that control the mutagenic properties of O6-meG. For example, we discovered that the C→T damage signature that arose in WT cells after MNNG treatment was present at a frequency of 7.5 × 10−5/bp (Figure 3A). Surprisingly though, the mutation frequency never exceeded 4.4 × 10−5/bp (Figure 4A), suggesting that only 60% of all misincorporation events result in bona fide mutations. One possibility is that the mismatch repair machinery is responsible for this discrepancy. When G:T mismatches are detected by the mismatch repair machinery in the absence of DNA replication, it cannot determine which base is correct and which base is mutant. As a result, it can only repair these mismatches with 50% accuracy (56), lowering the expected mutation frequency by approximately 50%.
Similar observations provide insight into the mutagenic properties of cells that lack the DNA repair protein mgt1p. In contrast to WT cells, dividing mgt1Δ cells convert 55% of lesions into mutations (Figure 4C), suggesting that only 45% of lesions are repaired over the course of 24 h. Although this conclusion seems to be contradicted by our measurements, which show that the frequency of O6-meG adducts returns to baseline levels after 24 h, it is important to remember that yeast cells create new, undamaged DNA strands during each round of cell division. As a result, the percentage of damaged bases could decline by as much as 50% every 90 min (the division rate of yeast cells) even in the absence of DNA repair. Thus, the lack of DNA repair in mgt1Δ cells is best demonstrated in arrested cells, which are not confounded by DNA replication (Figure 4D). Indeed, we found that in arrested mgt1Δ cells only 40% of lesions are repaired over a 24-h time span, closely matching the prediction made from our mutation measurements.
Even though we monitored O6-meG lesions after MNNG exposure to provide a proof of principle for our approach, it is likely that MADDD-seq can be used to detect other lesions as well. For example, we found that in addition to the G→A signature induced by O6-meG, arrested cells also displayed a distinct G→T damage signature. Although the molecular origin of this signal is currently unknown, one possible source of these lesions is 8-oxo-guanine. To determine if that is indeed the case, it will be important to validate MADDD-seq for additional lesions, including 8-oxo-guanine adducts. To do so, an analog approach could be used that is similar to the approach described here. For example, users interested in the detection of 8-oxo-guanine lesions could design oligos that carry this lesion at a specific location. These oligos can then be used to determine the mutagenic properties of 8-oxo-guanine during error-prone PCR, which will inform the number ‘family members’ that are required for damage detection. In addition, damaged and undamaged oligos can be used to optimize the PCR reactions to ensure efficient translesion synthesis, which is especially important if the lesion has a tendency to inhibit PCR amplification. Additional control reactions could be performed by mixing these oligos in different ratios, or adding them to extracted DNA samples, to determine the sensitivity of detection. Finally, treatment of naked DNA is recommended to ensure that damage is not mistaken for mutations during more complex experiments on living cells, while mutant cell lines will be an essential tool for final validation purposes, and the ratification of novel biological observations.
Care must also be given to the limitations of our assay. For example, even though MADDD-seq is extremely sensitive, it can only detect lesions that are mutagenic in nature. Our experiments with MNNG provide an important example of this limitation. Although the most important lesion created by MNNG is O6-meG, MNNG induces a range of other DNA lesions as well, none of which were detected by MADDD-seq because they are either not mutagenic at all, or not mutagenic enough to induce error-prone PCR during the first rounds of PCR amplification. In addition, it is important to note that MADDD-seq can only detect lesions that allow for efficient translesion synthesis. If the DNA polymerase used for PCR amplification is blocked by a lesion of interest, it cannot produce the full-length PCR amplicons required for detection. Potentially though, mutant DNA polymerases can be used to overcome this limitation. Finally, it is possible that DNA repair intermediates, mutagenic intermediates, or a combination of both can be mistaken for either damage or mutations during detection. For example, we found a C→T damage signature that is actually evidence of a mutation that is present in just one strand. Similarly, our data indicates that some O6-meG:T mismatches may have been reported as mutations.
One important advantage for end users is that MADDD-seq allows mutations and DNA damage to be detected simultaneously with a single assay, a single sample and a single reaction. Thus, DNA damage and mutations are always detected under identical conditions, thereby limiting unintended artifacts due to sample handling, batch effects, or user errors. Moreover, having a single assay for multiple endpoints means that precious materials such as human brain samples or tumor biopsies can be preserved for additional experiments in the future. Finally, simultaneous detection of multiple endpoints cuts back on the manpower, time and money that is needed to perform experiments, limits the expertise required to acquire usable data, and makes it easier to troubleshoot when results contradict each other. In doing so, we expect that MADDD-seq will prove to be a versatile, multi-functional detection tool for mutation research that can help basic researchers as well as clinical scientists reveal new biology that underlies human aging and disease.
Supplementary Material
Acknowledgements
We acknowledge support from the National Institute on Aging (R01AG054641 to M.V.), the University of Southern California (SCEHSC pilot award to M.V.), the National Institute of Environmental Health Sciences (U01ES029516 and R01ES026222 to J.H.B.), and the National Cancer Institute (R01CA204894 to J.H.B.).
Contributor Information
Marc Vermulst, University of Southern California, Leonard Davis School of Gerontology, Los Angeles, CA, USA.
Samantha L Paskvan, Translational Research Program, Public Health Sciences Division, Fred Hutchinson Cancer Research Center, Seattle, WA, USA.
Claire S Chung, University of Southern California, Leonard Davis School of Gerontology, Los Angeles, CA, USA.
Kathryn Franke, Translational Research Program, Public Health Sciences Division, Fred Hutchinson Cancer Research Center, Seattle, WA, USA.
Nigel Clegg, Translational Research Program, Public Health Sciences Division, Fred Hutchinson Cancer Research Center, Seattle, WA, USA.
Sam Minot, Data Core, Fred Hutchinson Cancer Research Center, Seattle, WA, USA.
Jennifer Madeoy, Translational Research Program, Public Health Sciences Division, Fred Hutchinson Cancer Research Center, Seattle, WA, USA.
Annalyssa S Long, Translational Research Program, Public Health Sciences Division, Fred Hutchinson Cancer Research Center, Seattle, WA, USA.
Jean-Francois Gout, Department of Biological Sciences, Mississippi State University, Mississippi State, MS, USA.
Jason H Bielas, Translational Research Program, Public Health Sciences Division, Fred Hutchinson Cancer Research Center, Seattle, WA, USA; Department of Laboratory Medicine and Pathology, University of Washington, Seattle, WA, USA.
Data availability
All sequencing data is freely available at SRA under accession number PRJNA1048550. Code to analyze MADDD-seq data is available at https://github.com/FredHutch/maddd-seq and https://doi.org/10.5281/zenodo.12636340.
Supplementary data
Supplementary Data are available at NAR Online.
Funding
National Institute on Aging [R01AG054641], [R01AG080365], [R01AG1075130] National Cancer Institute [R01CA204894]; National Institute of Environmental Health Sciences [R01ES026222, U01ES029516]; University of Southern California.
Conflict of interest statement. The Fred Hutchinson Cancer Research Center, of which JHB is a member, holds the patent for "duplex-sequencing technology". The remaining authors have no conflicts of interest to declare.
References
- 1. De Bont R., van Larebeke N.. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis. 2004; 19:169–185. [DOI] [PubMed] [Google Scholar]
- 2. Deman J., Van Larebeke N.. Carcinogenesis: mutations and mutagens. Tumour Biol. 2001; 22:191–202. [DOI] [PubMed] [Google Scholar]
- 3. Lindahl T. Instability and decay of the primary structure of DNA. Nature. 1993; 362:709–715. [DOI] [PubMed] [Google Scholar]
- 4. Hu J., Lieb J.D., Sancar A., Adar S.. Cisplatin DNA damage and repair maps of the human genome at single-nucleotide resolution. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:11507–11512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Shu X., Xiong X., Song J., He C., Yi C.. Base-resolution analysis of cisplatin-DNA adducts at the genome scale. Angew. Chem. Int. Ed. Engl. 2016; 55:14246–14249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Hu J., Li W., Adebali O., Yang Y., Oztas O., Selby C.P., Sancar A.. Genome-wide mapping of nucleotide excision repair with XR-seq. Nat. Protoc. 2019; 14:248–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li W., Hu J., Adebali O., Adar S., Yang Y., Chiou Y.Y., Sancar A.. Human genome-wide repair map of DNA damage caused by the cigarette smoke carcinogen benzo[a]pyrene. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:6752–6757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Hu J., Adebali O., Adar S., Sancar A.. Dynamic maps of UV damage formation and repair for the human genome. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:6758–6763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Friedberg E.C., Walker G.C., Siede W., Wood R.D., Schultz R.A., Ellenberger T.. DNA Repair and Mutagenesis. 2006; 2 edWashington DC: American Society for Microbiology. [Google Scholar]
- 10. Sloan D.B., Broz A.K., Sharbrough J., Wu Z.. Detecting rare mutations and DNA damage with sequencing-based methods. Trends Biotechnol. 2018; 36:729–740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Mingard C., Wu J., McKeague M., Sturla S.J.. Next-generation DNA damage sequencing. Chem. Soc. Rev. 2020; 49:7354–7377. [DOI] [PubMed] [Google Scholar]
- 12. Gregory M.T., Bertout J.A., Ericson N.G., Taylor S.D., Mukherjee R., Robins H.S., Drescher C.W., Bielas J.H.. Targeted single molecule mutation detection with massively parallel sequencing. Nucleic Acids Res. 2016; 44:e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wyatt M.D., Pittman D.L.. Methylating agents and DNA repair responses: methylated bases and sources of strand breaks. Chem. Res. Toxicol. 2006; 19:1580–1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Goodwin S., McPherson J.D., McCombie W.R.. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016; 17:333–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rehm H.L. Evolving health care through personal genomics. Nat. Rev. Genet. 2017; 18:259–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Alexandrov L.B., Kim J., Haradhvala N.J., Huang M.N., Tian Ng A.W., Wu Y., Boot A., Covington K.R., Gordenin D.A., Bergstrom E.N.et al.. The repertoire of mutational signatures in human cancer. Nature. 2020; 578:94–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kennedy S.R., Salk J.J., Schmitt M.W., Loeb L.A.. Ultra-sensitive sequencing reveals an age-related increase in somatic mitochondrial mutations that are inconsistent with oxidative damage. PLoS Genet. 2013; 9:e1003794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Rosendahl Huber A., Van Hoeck A., Van Boxtel R. The mutagenic impact of environmental exposures in Human cells and cancer: imprints through time. Front. Genet. 2021; 12:760039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kennedy S.R., Schmitt M.W., Fox E.J., Kohrn B.F., Salk J.J., Ahn E.H., Prindle M.J., Kuong K.J., Shen J.C., Risques R.A.et al.. Detecting ultralow-frequency mutations by Duplex sequencing. Nat. Protoc. 2014; 9:2586–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Schmitt M.W., Kennedy S.R., Salk J.J., Fox E.J., Hiatt J.B., Loeb L.A.. Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:14508–14513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Salk J.J., Schmitt M.W., Loeb L.A.. Enhancing the accuracy of next-generation sequencing for detecting rare and subclonal mutations. Nat. Rev. Genet. 2018; 19:269–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Margison G.P., Santibanez Koref M.F., Povey A.C.. Mechanisms of carcinogenicity/chemotherapy by O6-methylguanine. Mutagenesis. 2002; 17:483–487. [DOI] [PubMed] [Google Scholar]
- 23. Cancer Genome Atlas Research, N Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008; 455:1061–1068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Chung J., Das A., Sun X., Sobreira D.R., Leung Y.Y., Igartua C., Mozaffari S., Chou Y.F., Thiagalingam S., Mez J.et al.. Genome-wide association and multi-omics studies identify MGMT as a novel risk gene for Alzheimer's disease among women. Alzheimers Dement. 2022; 10.1002/alz.12719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Bielas J.H., Heddle J.A.. Proliferation is necessary for both repair and mutation in transgenic mouse cells. Proc. Natl. Acad. Sci. U.S.A. 2000; 97:11391–11396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Yaakoub H., Mina S., Calenda A., Bouchara J.P., Papon N.. Oxidative stress response pathways in fungi. Cell. Mol. Life Sci. 2022; 79:333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang D., Kreutzer D.A., Essigmann J.M.. Mutagenicity and repair of oxidative DNA damage: insights from studies using defined lesions. Mutat. Res. 1998; 400:99–115. [DOI] [PubMed] [Google Scholar]
- 28. Sharma S., Baysal B.E.. Stem-loop structure preference for site-specific RNA editing by APOBEC3A and APOBEC3G. PeerJ. 2017; 5:e4136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Oman M., Alam A., Ness R.W.. How sequence context-dependent mutability drives mutation rate variation in the genome. Genome Biol. Evol. 2022; 14:evac032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lou D.I., Hussmann J.A., McBee R.M., Acevedo A., Andino R., Press W.H., Sawyer S.L.. High-throughput DNA sequencing errors are reduced by orders of magnitude using circle sequencing. Proc. Natl. Acad. Sci. U.S.A. 2013; 110:19872–19877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang K., Lai S., Yang X., Zhu T., Lu X., Wu C.I., Ruan J.. Ultrasensitive and high-efficiency screen of de novo low-frequency mutations by o2n-seq. Nat. Commun. 2017; 8:15335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Maslov A.Y., Makhortov S., Sun S., Heid J., Dong X., Lee M., Vijg J.. Single-molecule, quantitative detection of low-abundance somatic mutations by high-throughput sequencing. Sci. Adv. 2022; 8:eabm3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zatopek K.M., Potapov V., Maduzia L.L., Alpaslan E., Chen L., Evans T.C., Ong J.L., Ettwiller L.M., Gardner A.F. RADAR-seq: a RAre DAmage and repair sequencing method for detecting DNA damage on a genome-wide scale. DNA Repair (Amst.). 2019; 80:36–44. [DOI] [PubMed] [Google Scholar]
- 34. Hwa Yun B., Guo J., Bellamri M., Turesky R.J. DNA adducts: formation, biological effects, and new biospecimens for mass spectrometric measurements in humans. Mass Spectrom. Rev. 2020; 39:55–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Phillips D.H., Arlt V.M.. (32)P-postlabeling analysis of DNA adducts. Methods Mol. Biol. 2020; 2102:291–302. [DOI] [PubMed] [Google Scholar]
- 36. Gorini F., Scala G., Di Palo G., Dellino G.I., Cocozza S., Pelicci P.G., Lania L., Majello B., Amente S.. The genomic landscape of 8-oxodG reveals enrichment at specific inherently fragile promoters. Nucleic Acids Res. 2020; 48:4309–4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Fang Y., Zou P.. Genome-wide mapping of oxidative DNA damage via engineering of 8-oxoguanine DNA glycosylase. Biochemistry. 2020; 59:85–89. [DOI] [PubMed] [Google Scholar]
- 38. Poetsch A.R., Boulton S.J., Luscombe N.M.. Genomic landscape of oxidative DNA damage and repair reveals regioselective protection from mutagenesis. Genome Biol. 2018; 19:215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mao P., Brown A.J., Malc E.P., Mieczkowski P.A., Smerdon M.J., Roberts S.A., Wyrick J.J.. Genome-wide maps of alkylation damage, repair, and mutagenesis in yeast reveal mechanisms of mutational heterogeneity. Genome Res. 2017; 27:1674–1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mao P., Smerdon M.J., Roberts S.A., Wyrick J.J.. Chromosomal landscape of UV damage formation and repair at single-nucleotide resolution. Proc. Natl. Acad. Sci. U.S.A. 2016; 113:9057–9062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Baranello L., Kouzine F., Wojtowicz D., Cui K., Przytycka T.M., Zhao K., Levens D.. DNA break mapping reveals topoisomerase II activity genome-wide. Int. J. Mol. Sci. 2014; 15:13111–13122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Sriramachandran A.M., Petrosino G., Mendez-Lago M., Schafer A.J., Batista-Nascimento L.S., Zilio N., Ulrich H.D.. Genome-wide nucleotide-resolution mapping of DNA replication patterns, single-strand breaks, and lesions by GLOE-seq. Mol. Cell. 2020; 78:975–985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Crosetto N., Mitra A., Silva M.J., Bienko M., Dojer N., Wang Q., Karaca E., Chiarle R., Skrzypczak M., Ginalski K.et al.. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat. Methods. 2013; 10:361–365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Canela A., Sridharan S., Sciascia N., Tubbs A., Meltzer P., Sleckman B.P., Nussenzweig A.. DNA breaks and end resection measured genome-wide by end sequencing. Mol. Cell. 2016; 63:898–911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Koh K.D., Balachander S., Hesselberth J.R., Storici F.. Ribose-seq: global mapping of ribonucleotides embedded in genomic DNA. Nat. Methods. 2015; 12:251–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Clausen A.R., Lujan S.A., Burkholder A.B., Orebaugh C.D., Williams J.S., Clausen M.F., Malc E.P., Mieczkowski P.A., Fargo D.C., Smith D.J.et al.. Tracking replication enzymology in vivo by genome-wide mapping of ribonucleotide incorporation. Nat. Struct. Mol. Biol. 2015; 22:185–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Shu X., Liu M., Lu Z., Zhu C., Meng H., Huang S., Zhang X., Yi C.. Genome-wide mapping reveals that deoxyuridine is enriched in the human centromeric DNA. Nat. Chem. Biol. 2018; 14:680–687. [DOI] [PubMed] [Google Scholar]
- 48. Li S., Waters R., Smerdon M.J.. Low- and high-resolution mapping of DNA damage at specific sites. Methods. 2000; 22:170–179. [DOI] [PubMed] [Google Scholar]
- 49. Besaratinia A., Pfeifer G.P.. Measuring the formation and repair of UV damage at the DNA sequence level by ligation-mediated PCR. Methods Mol. Biol. 2012; 920:189–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lehle S., Hildebrand D.G., Merz B., Malak P.N., Becker M.S., Schmezer P., Essmann F., Schulze-Osthoff K., Rothfuss O.. LORD-Q: a long-run real-time PCR-based DNA-damage quantification method for nuclear and mitochondrial genome analysis. Nucleic Acids Res. 2014; 42:e41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Hu J., Adar S.. The cartography of UV-induced DNA damage formation and DNA repair. Photochem. Photobiol. 2017; 93:199–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Valente W.J., Ericson N.G., Long A.S., White P.A., Marchetti F., Bielas J.H.. Mitochondrial DNA exhibits resistance to induced point and deletion mutations. Nucleic Acids Res. 2016; 44:8513–8524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Teuber-Hanselmann S., Worm K., Macha N., Junker A.. MGMT-methylation in non-neoplastic diseases of the Central nervous system. Int. J. Mol. Sci. 2021; 22:3845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Gerson S.L. MGMT: its role in cancer aetiology and cancer therapeutics. Nat. Rev. Cancer. 2004; 4:296–307. [DOI] [PubMed] [Google Scholar]
- 55. Miller M.B., Huang A.Y., Kim J., Zhou Z., Kirkham S.L., Maury E.A., Ziegenfuss J.S., Reed H.C., Neil J.E., Rento L.et al.. Somatic genomic changes in single Alzheimer's disease neurons. Nature. 2022; 604:714–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Rodriguez G.P., Romanova N.V., Bao G., Rouf N.C., Kow Y.W., Crouse G.F.. Mismatch repair-dependent mutagenesis in nondividing cells. Proc. Natl. Acad. Sci. U.S.A. 2012; 109:6153–6158. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data is freely available at SRA under accession number PRJNA1048550. Code to analyze MADDD-seq data is available at https://github.com/FredHutch/maddd-seq and https://doi.org/10.5281/zenodo.12636340.