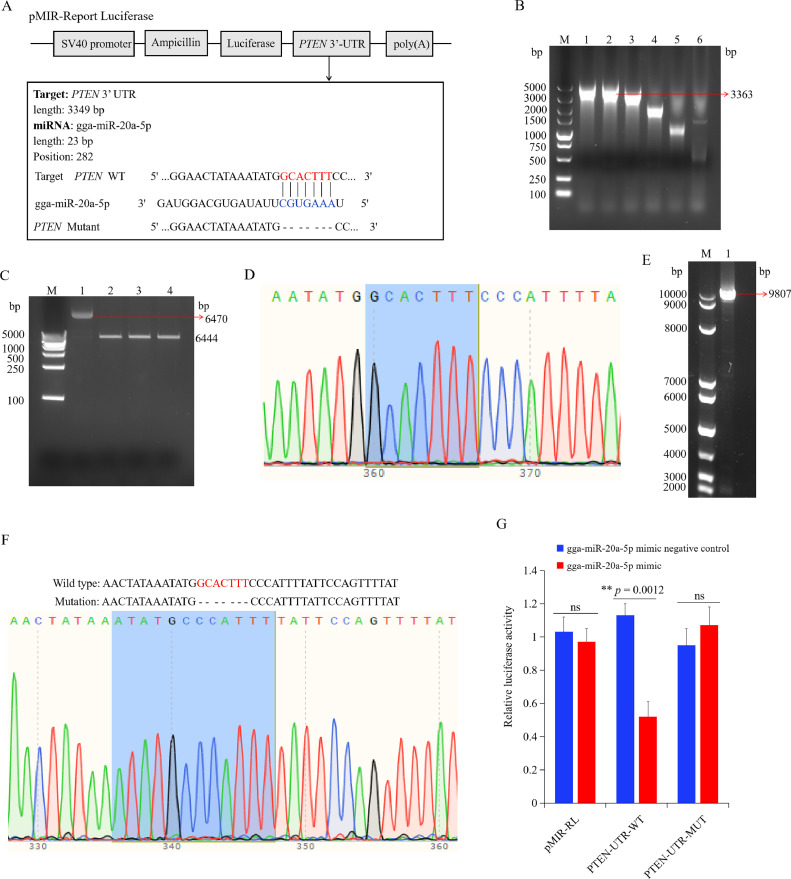
Figure 9.
PTEN was a directly target of gga-miR-20a-5p. (A) The sequence alignment of gga-miR-20a-5p and 1 target site in the 3′ UTR of PTEN is shown. (B) The PCR product of wild type 3′ UTR of PTEN with different values of Tm. M, Marker; 1 to 6, wild type 3′ UTR of PTEN production with Tm of 55°C, 57°C, 59°C, 61°C, 63°C, 65°C. (C) Double enzyme digestion of the pMIR-report luciferase plasmid. M, Marker; 1, circular pMIR-Report Luciferase plasmid; 2 to 4, the linear pMIR-report luciferase plasmid after double enzyme digestion. (D) The sequencing result of wild type 3′ UTR of PTEN. (E) Construction of the mutant of PTEN 3′ UTR. M, Marker; 1, The PCR product amplified by the mutant 3′UTR of PTEN primers using the wild type 3′UTR of PTEN plasmid as a template. (F) The sequencing result of mutant 3′UTR of PTEN plasmid. (G) Chicken macrophages were co-transfected with pMIR-report luciferase or 3′UTR of PTEN luciferase reporter plasmid (wild-type or mutation), along with mimics negative control or gga-miR-20a-5p mimics as indicated. Firefly luciferase activity was measured and normalized using renilla luciferase activity to finally calculate the relative luciferase activity 48 h after transfection. Data are shown as mean ± SD; n = 4 independent experiments; paired t test; ns, not significant; **P < 0.01.