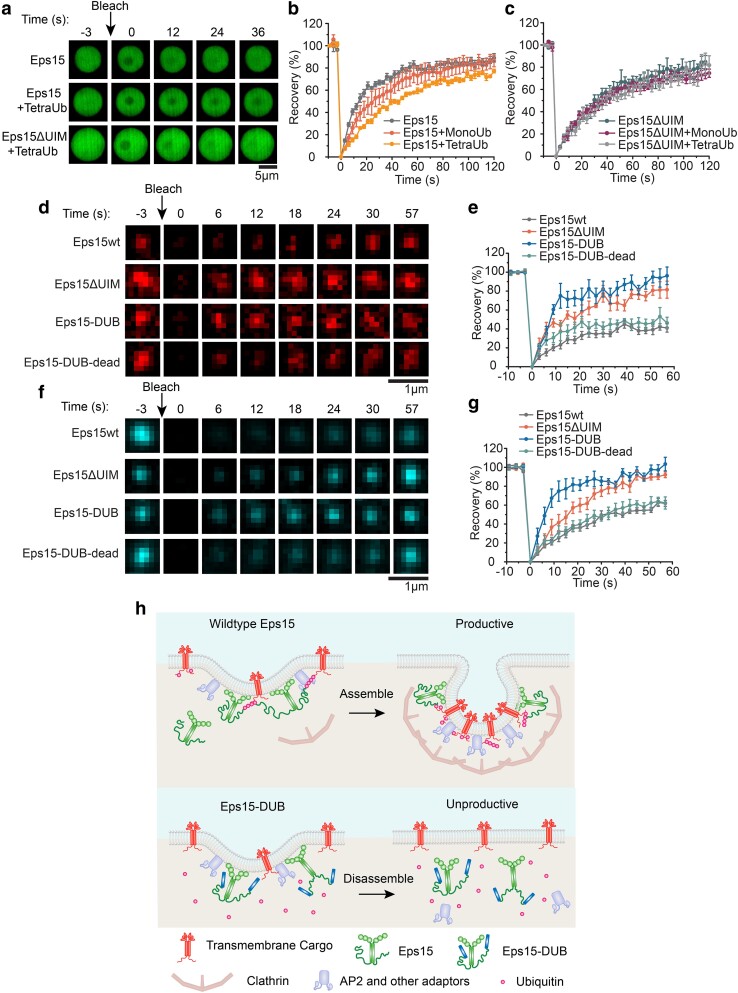
Fig. 5.
In condensates and at endocytic sites, loss of Eps15–ubiquitin interactions results in increased molecular exchange of Eps15. a) Representative image series of FRAP of Eps15, Eps15 + TetraUb, and Eps15ΔUIM + TetraUb droplets, respectively. Scale bar = 5 μm. b, c) Fluorescence recovery curves for Eps15 (b) or Eps15ΔUIM (c) droplets in the presence of MonoUb or TetraUb. Eps15/Eps15ΔUIM concentration was maintained at 7 μM with the addition of 1 μM MonoUb and 0.25 μM TetraUb, respectively. Droplets were made in 20 mM Tris–HCl, 150 mM NaCl, 5 mM TCEP, 1 mM EDTA, and 1 mM EGTA at pH 7.5 with 3% w/v PEG8000 and all droplet experiments were conducted at room temperature. Data shown as mean ± SE. n = 6 droplets under each condition. Mobile fractions and t1/2: Eps15 (85.9 ± 5.3%, 12.5 ± 0.8 s), Eps15 + MonoUb (82.7 ± 10.9%, 18.0 ± 1.3 s), Eps15 + TetraUb (79.0 ± 9.3%, 31.1 ± 1.2 s), Eps15ΔUIM (81.5 ± 12.1%, 21.4 ± 1.1 s), Eps15ΔUIM + MonoUb (75.74 ± 6.5%, 21.8 ± 1.9 s), and Eps15ΔUIM + TetraUb (76.3 ± 11.3%, 24.12 ± 1.6 s). d, e) Representative images of fluorescence recovery of Eps15-mCherry variants in clathrin-coated structures in Eps15KO cells expressing corresponding variants (d) and the average fluorescence recovery plots for each condition (e). Mobile fractions and t1/2: Eps15wt (41.3%, 19.1 s), Eps15ΔUIM (81.5%, 5.5 s), Eps15-DUB (96.7%, 8.0 s), and Eps15-DUB-dead (46.3%, 11.5 s). f, g) Representative images of fluorescence recovery of AP2-HaloTag labeled with JF646 in clathrin-coated structures in Eps15KO cells expressing corresponding Eps15 variants (f) and the average fluorescence recovery plots (g). Mobile fractions and t1/2: Eps15wt (71.9%, 29.7 s), Eps15ΔUIM (100.0%, 15.1 s), Eps15-DUB (103.9%, 6.8 s), and Eps15-DUB-dead (66.6%, 22.9 s). n = 6 pits were analyzed for each plot. Data were shown as mean ± standard error. Scale bar = 1 μm. All cell FRAP experiments were performed at 37°C. h) Schematic showing how polyubiquitin stabilizes the endocytic protein network by interacting with and cross-linking UIMs on endocytic proteins, resulting in productive clathrin-mediated endocytosis (Top). Removal of ubiquitin from the endocytic protein network using DUB decreases the network multivalency thus making the network less stable, resulting in less efficient clathrin-mediated endocytosis (Bottom).