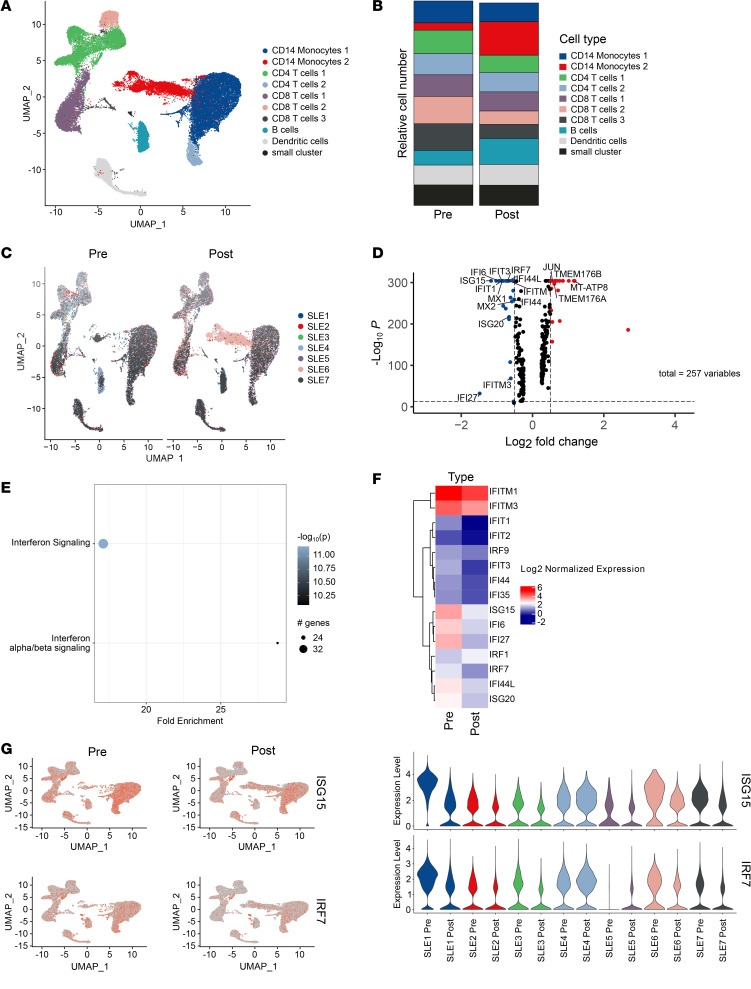
Figure 1. scRNA-Seq–based analysis of PBMCs of patients with SLE.
(A–G) Single-cell RNA-Seq–based (scRNA-Seq–based) analysis of peripheral blood mononuclear cells (PBMCs) of patients with SLE (n = 7) before CD19 CAR T cell therapy and after early B cell repopulation. Single data sets from each patient were integrated using the Seurat pipeline into one data set containing approximately 40,700 PBMC. (A) Dimensional reduction of PBMCs revealing indicated UMAP clusters where cluster identities were determined through top cluster gene analysis and the Enrichr package. (B and C) Comparison of single-cell data sets from patients with SLE before anti-CD19 CAR T cell therapy and after early B cell reconstitution. (D) Differential gene expression analysis on the effect of anti-CD19 CAR T cell therapy. (E) Pathway enrichment analysis based on changes in gene expression of total PBMCs. The color intensity reflects the adjusted P value for each pathway. (F) IFN-associated genes are highlighted in a normalized heatmap representing mean gene expression of interferon related genes (n = 15 unique genes) before and after CD19 CAR T cell treatment. (G) Gene expression levels of ISG15 and IRF7 as representative IFN-induced genes are highlighted as a feature plot and in violin plots illustrating expression levels on the basis of individual patients.