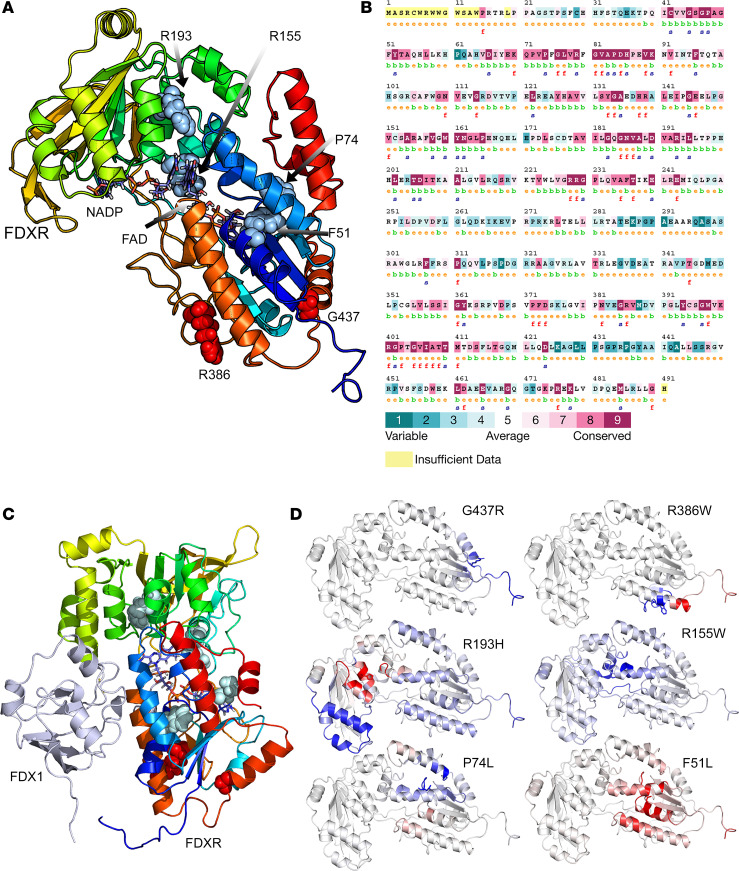
Figure 3. Sequence and structure analysis of mutations in FDXR.
(A) A 3D model of human FDXR displayed as a ribbon diagram. The positions of the phenylalanine 51, proline 74, arginine 155, arginine 193, arginine 386, and glycine 437 residues are indicated. The structural model of human FDXR is based on a known 3D structure of the bovine protein as described in the Methods section. The diagram is colored using a rainbow palette with blue at N-terminus and red at C-terminus. Cofactors (NADP, FAD) are shown as stick models, while amino acids phenylalanine 51, proline 74, arginine 155, arginine 193, arginine 386, and glycine 437 are shown as spheres. (B) The evolutionary sequence conservation of FDXR. Most of the mutations reported in this study are highly conserved across species and are predicted to have structural roles. Sequences are colored based on amino acid conservation, with dark blue being the least conserved and dark red being the most conserved, while yellow indicates that no prediction could be made. (C) A complex of FDXR and FDX1 proteins showing the locations of mutated residues, which are not at the FDXR-FDX interface and are predicted not to have a direct effect on FDX-FDXR interaction. (D) Stability and flexibility analysis of mutated FDXR structures compared with WT FDXR. An increased flexibility was observed for amino acid changes F51L and R193H (shown in red), indicating decreased stability that was supported by differential free energy calculations. Decreased flexibility due to P74L, R155W, R386W, and G537R mutations is shown in blue.