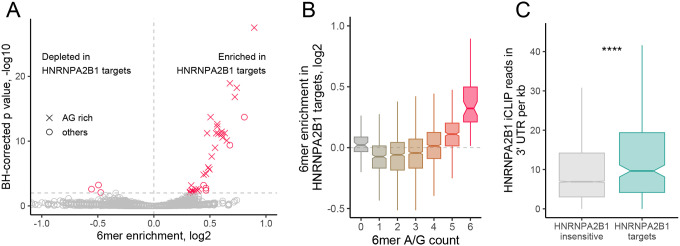
Figure 3.
HNRNPA2B1 localization targets are enriched for HNRNPA2B1 motifs and CLIP sites in their 3′ UTRs. (A) Enrichments for all 6mers in the 3′ UTRs of HNRNPA2B1 targets RNAs compared to the 3′ UTRs of RNAs of HNRNPA2B1-insensitive RNAs. 6mers that are completely comprised of A and G are represented with X. All others are represented with circles. (B) Distributions of 6mer enrichments in the 3′ UTRs of HNRNPA2B1 targets RNAs compared to the 3′ UTRs of RNAs of HNRNPA2B1-insensitive RNAs. 6mers are binned according to the number of A/G they contain. (C) HNRNPA2B1 CLIP read densities in the 3′ UTRs of HNRNPA2B1-insensitive (gray) and HNRNPA2B1-target (teal) RNAs. P-values were calculated using a Wilcoxon ranksum test. NS (not significant) represents p > 0.05, * p < 0.05, ** p < 0.01, *** p < 0.001, and **** represents p < 0.0001.