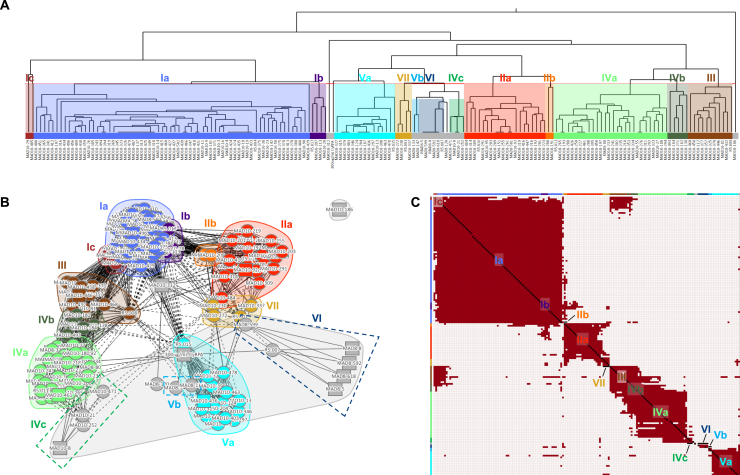
Figure S2.
Detailed epitope binning analysis based on mAb competition profiles, related to Figure 3
(A) Hierarchical clustering of mAbs using the McQuitty method (Epitope software), based on SPR-based mAb competition. Epitope bins were derived from mAb clustering and competition profiles and are shown in different colors. Bins Ia–Vb were identified based on the hierarchical tree, with two mAbs in Ic clustered into a separate bin due to limited overlap with group Ia mAbs and extensive competition with group III mAbs (see Figure S2C, top left). Bin VI was identified based on competition with the anchor mAb R5.007, which targets the RH5 intrinsic loop.
(B) Epitope bins of RH5-specific mAbs shown in a two-dimensional (2D) representation, clustered based on competition profiles. Solid lines indicate two-way competition, dashed lines indicate one-way competition. Circular nodes represent mAbs analyzed as both ligand and analyte; square nodes represent mAbs analyzed as ligand or analyte only. The layout.auto display (Epitope software) was used to generate this plot.
(C) Heatmap showing competition of mAbs targeting RH5. Red squares signify competition between the corresponding analyte and ligand mAb, while cream squares indicate no competition. The colors at the side and top are those of the 13 individual bins. 13/22 infection-derived mAbs and 153/164 vaccine-derived mAbs were mappable in this assay; the remaining mAbs were incompatible with the workflow, e.g., due to acid sensitivity during the regeneration step.