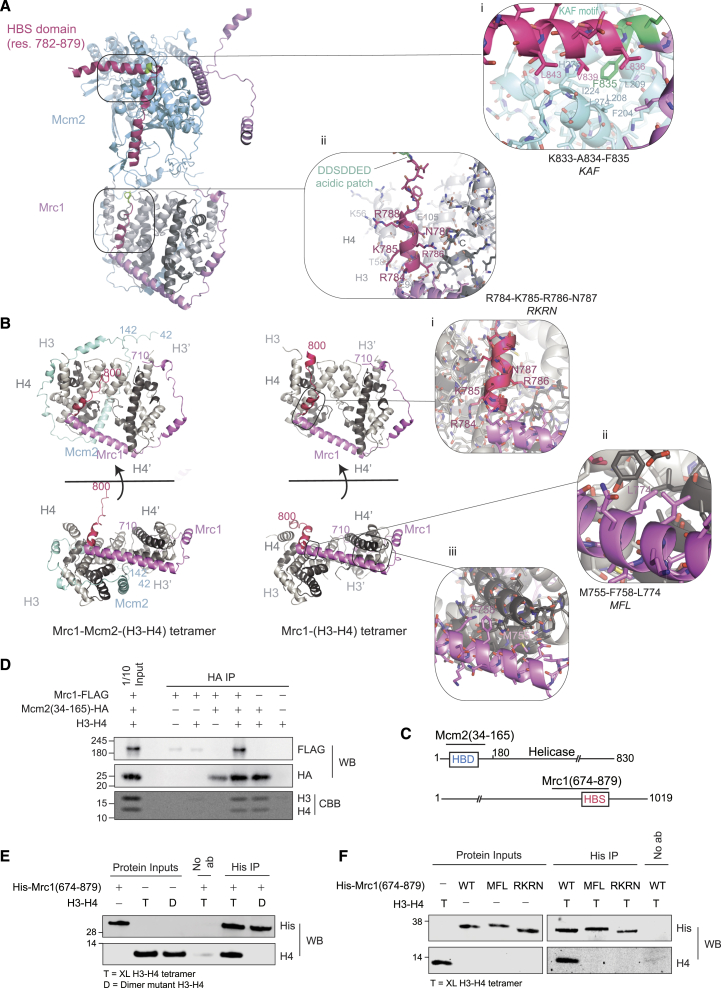
Figure 5.
Mrc1 chaperones H3-H4 tetramers in a manner compatible with Mcm2 co-chaperoning
(A) AF90,91 prediction of a complex comprising full-length S. pombe Mrc1 (pink) and Mcm2 (light blue) bound to a histone H3-H4 tetramer (gray). Histone tails and unstructured Mrc1 residues predicted with low confidence (residues 1–381, 475–710, 800–804, and 856–1,020) are not depicted for clarity. Closeups: (1) interaction of the Mrc1 KAF motif with a hydrophobic groove in Mcm2 and (2) interaction of the N-terminal alpha helix of the Mrc1 HBS domain with charged aa in histone H3 αN and α2 helices and histone H4 C terminus.
(B) AF predictions of Mrc1 histone binding via the HBD, including beginning of the HBS domain (dark pink) and upstream region (710–800). Interaction between Mrc1 (pink) and the histones (gray) remains unchanged in the presence of Mcm2 (light blue). Closeups highlight residues subjected to mutational analysis.
(C) Mcm2(34–165) and Mrc1(674–879) polypeptides used for pull-downs.
(D) Pull-downs of full-length Mrc1-FLAG and H3-H4 with HA-Mcm2 HBD (34–165). Pulled-down proteins were detected by western blot (WB) or Coomassie staining (CBB). MW in KDa.
(E) Pull-downs of H3-H4 dimers or tetramers with His6-Mrc1(674–879).
(F) Pull-downs of H3-H4 tetramers with His6-Mrc1(674–879) WT and indicated mutants.