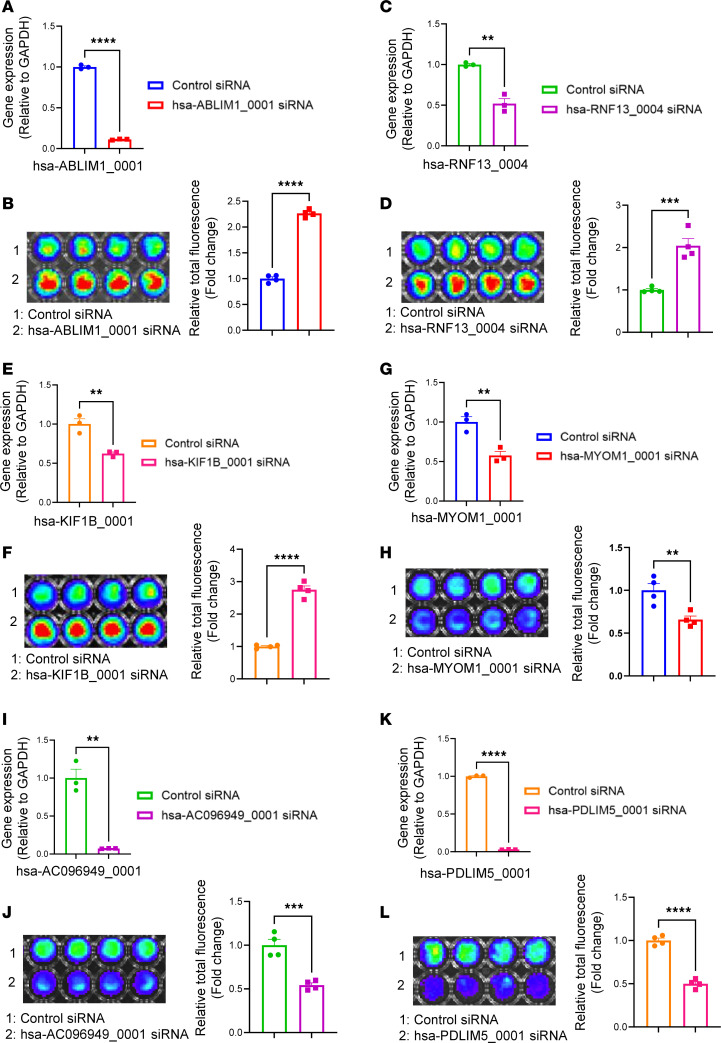
Figure 5. circRNAs that modulate cardiomyocyte cell cycle.
hiPSC-CMs at day 28 after initiation of cardiac differentiation were utilized. The efficiency of siRNA-based knockdown of circRNAs in hiPSC-CMs was determined by qRT-PCR. The relative expression levels of circRNAs were normalized to GAPDH reference gene. Cell number was evaluated by bioluminescence analysis. Data were presented as percentage. (A, C, E, G, I, and K) Evaluation of the expression of hsa-ABLIM1_0001, hsa-RNF13_0004, hsa-KIF1B_0001, hsa-MYOM1_0001, hsa-AC096949_0001, and hsa-PDLIM5_0001 in hiPSC-CMs after siRNA treatment. (B, D, F, H, J, and L) Bioluminescence assay and quantification of bioluminescent signal intensity in hiPSC-CMs with siRNA-based knockdown of the specific circRNAs. All data were presented as mean ± SEM. Statistical analysis was performed via 2-tailed Student’s t test. n = 3 technical replicates in each group for panels A, C, E, G, I, and K. n = 4 technical replicates in each group for panels B, D, F, H, J, and L. **P < 0.01, ***P < 0.001, and ****P < 0.0001.