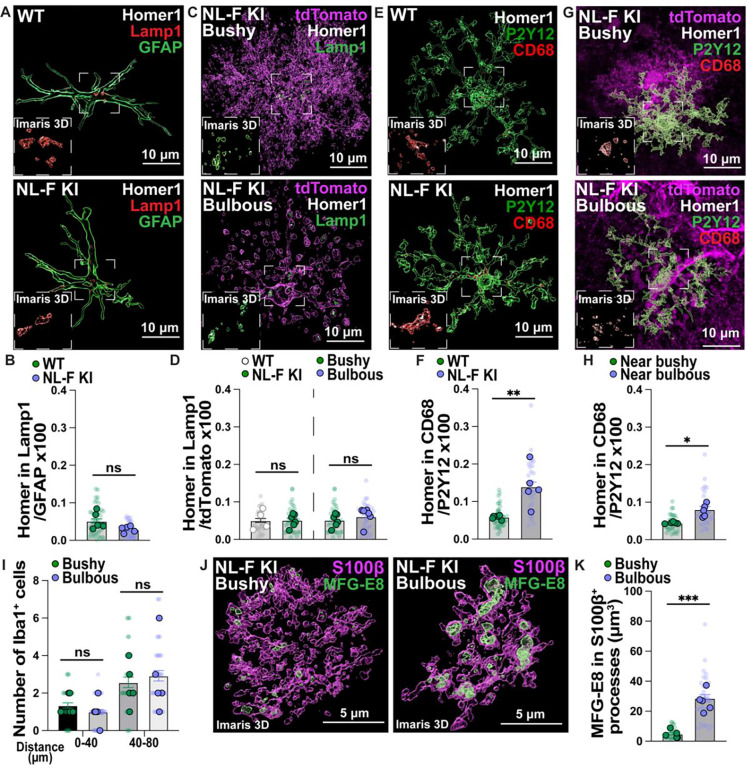
Fig 3. Increased microglia-Homer1 engulfment near hippocampal NL-F KI bulbous astrocytes.
A) Representative 3D rendered images of excitatory post-synaptic Homer1 (white), Lamp1 lysosomes (red) and GFAP (green) in the 6-mo hippocampal CA1 SR. Scale bar = 10 μm. Inset shows representative zoom of Homer1 inside Lamp1+ astrocytic lysosomes. B) Quantification of astrocytic Homer1 engulfment using Imaris 3D surface rendering shown as: Homer1 volume in Lamp1+ lysosomes in GFAP+ astrocytic surface/GFAP volume x100. Transparent points = individual astrocytes (5–10 astrocytes per mouse; 40 WT and 39 NL-F KI astrocytes sampled in total); full points = mouse average of ROIs (n=5 male mice per genotype). Unpaired two-tailed student’s t-test with Welch’s correction on mouse average. C) Representative 3D rendered images of excitatory post-synaptic Homer1 (white), Lamp1 lysosomes (green) and tdTomato (magenta) in the 6-mo hippocampal CA1 SR. Scale bar = 10 μm. Inset shows representative zoom of Homer1 inside Lamp1+ astrocytic lysosomes. D) Quantification of astrocytic Homer1 engulfment using Imaris 3D surface rendering shown as: Homer1 volume in Lamp1+ lysosomes in tdTomato+ astrocytic surface/tdTomato volume x100. Transparent points = individual astrocytes (4–8 astrocytes per condition; 26 WT, 35 NL-F KI bushy and 31 NL-F KI bulbous astrocytes sampled in total), full points = mouse average of ROIs (n=4–6 male mice). Unpaired (WT vs NL-F KI) or paired (NL-F KI bushy vs NL-F KI bulbous) two-tailed student’s t-test on mouse average. Dashed line represents graph split. E) Representative 3D rendered images of excitatory post-synaptic Homer1 (white), CD68 lysosomes (red) and P2Y12 (green) in the 6-mo hippocampal CA1 SR. Scale bar = 10 μm. Inset shows representative zoom of Homer1 inside CD68+ microglial lysosomes. F) Quantification of microglial Homer1 engulfment using Imaris 3D surface rendering shown as: Homer1 volume in CD68+ lysosomes in P2Y12+ microglial surface/P2Y12 volume x100. Transparent points = individual microglia (5–6 microglia per mouse; 36 WT and 28 NL-F KI microglia sampled in total); full points = mouse average of ROIs (n=5–6 male mice per genotype). Two-tailed Mann Whitney test on mouse average. G) Representative 3D rendered images of excitatory post-synaptic Homer1 (white), CD68 lysosomes (red) and P2Y12 (green) in the 6-mo hippocampal CA1 SR near bushy or bulbous tdTomato+ (magenta) astrocytes. Scale bar = 10 μm. Inset shows representative zoom of Homer1 inside CD68+ microglial lysosomes. H) Quantification of microglial Homer1 engulfment using Imaris 3D surface rendering shown as: Homer1 volume in CD68+ lysosomes in P2Y12+ microglial surface/P2Y12 volume x100. Transparent point = individual microglia (5–7 microglia per condition; 32 and 28 microglia sampled in total near (< 5 μm away) bushy and bulbous NL-F KI astrocytes respectively), full points = mouse average of ROIs (n=6 male mice). Paired two-tailed student’s t-test on mouse average. I) Quantification of the number of Iba1+ microglia near bushy or bulbous NL-F KI astrocytes. Transparent points = individual astrocyte regions (4–10 regions per condition; 30 NL-F KI bushy and 33 NL-F KI bulbous astrocyte regions sampled in total); full points = mouse average of ROIs (n=5 male mice). Wilcoxon matched-pairs signed rank test followed by a Bonferroni-Dunn correction on mouse average. J) Representative 3D rendered images of MFG-E8 (green) inside S100β+ (magenta) bushy or bulbous astrocytic processes in the 6-mo NL-F KI hippocampus. Scale bar = 5 μm. K) Quantification of the volume of MFG-E8 within bushy or bulbous S100β+ astrocytic processes per μm3 using Imaris 3D surface rendering. Transparent points = individual astrocytes (3–5 astrocytes per condition; 27 NL-F KI bushy and 30 NL-F KI bulbous astrocytes sampled in total); full points = mouse average of ROIs (n=5 male mice). Paired two-tailed student’s t-test on mouse average. All data shown as mean ± SEM. p-values shown ns P>0.05; *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.