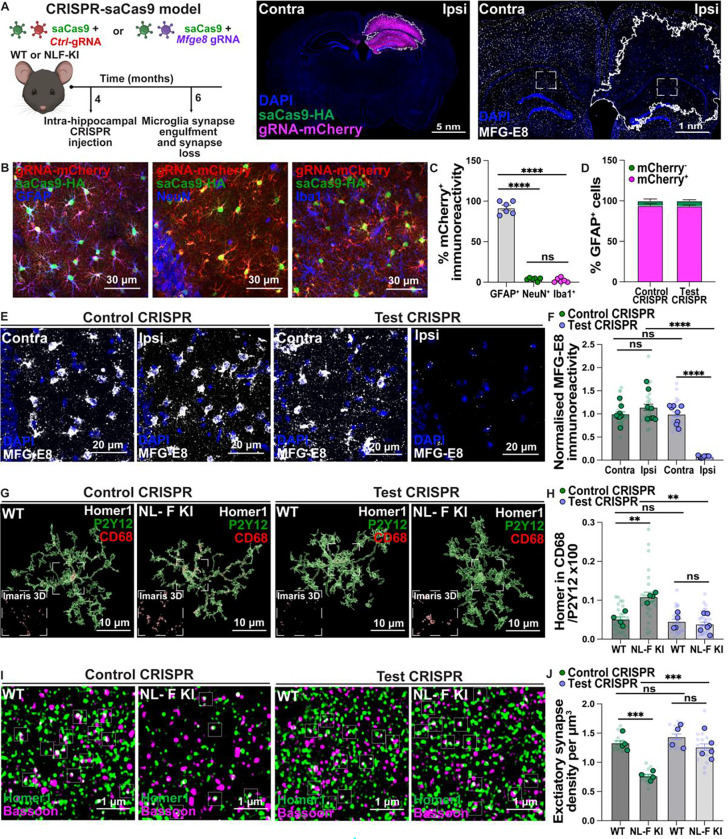
Fig 5. Viral CRISPR-saCas9 deletion of astrocytic MFG-E8 prevents microglia-Homer1 engulfment and excitatory loss in the NL-F KI hippocampus.
A) Schematic of CRISPR-saCas9 paradigm to knock-down Mfge8 from hippocampal astrocytes with spatio-temporal control. Immunostaining for gRNA-mCherry (magenta) and Cas9-HA (green) constructs post-CRISPR-saCas9 injection. Scale bar = 5 nm. Representative image of MFG-E8 (white) immunoreactivity post-CRISPR-saCas9 injection. Scale bar = 1 nm. Inset highlights ROIs. mCherry injection zone drawn with white ROI on ImageJ. B) Representative images of gRNA-mCherry (red) and Cas9-HA (green) constructs with either GFAP (blue, left), NeuN (blue, middle) or Iba1 (blue, right) post-CRISPR-saCas9 injection. Scale bar = 30 μm. C) Quantification of the colocalized mCherry-labeled signal with GFAP+ astrocytes, NeuN+ neurons or Iba1+ microglia in the hippocampus using ImageJ shown as percentage of the total mCherry-labeled signal. Each point = 1 mouse (n=6 mixed male and female mice). Brown-Forsythe (643.8 (2.000, 6.920, p<0.0001) and Welch ANOVA (339.9 (2.000, 8.829), p<0.0001) with Dunnett’s T3 multiple comparisons test. D) Quantification of the colocalized GFAP+ astrocytes with mCherry-labelled signal in the hippocampus using ImageJ shown as percentage of the total GFAP+ astrocytes. 8–10 male mice sampled. E) Representative images of MFG-E8 (white) immunoreactivity in the 6-mo WT and NL-F KI SR hippocampus post control or test CRISPR-saCas9 injection. Scale bar = 20 μm. F) Quantification of MFG-E8 immunoreactivity using ImageJ. Data shown as normalized to contra control CRISPR injected hemisphere. Transparent points = individual ROIs (3 ROIs per condition; 24 ROIs sampled in total per condition), full points = mouse average of ROIs (n=8 male mice per condition). Two-way ANOVA (Interaction: F (1, 28) = 218.6, p<0.0001) followed by Bonferroni’s multiple comparisons post-hoc test on Y=log(Y) transformed mouse average. G) Representative 3D rendered images of excitatory post-synaptic Homer1 (white), CD68 lysosomes (red) and P2Y12 (green) in the 6-mo WT and NL-F KI CA1 SR hippocampus post control or test CRISPR-saCas9 injection. Scale bar = 10 μm. Inset shows representative zoom of Homer1 inside CD68+ microglial lysosomes. H) Quantification of microglial Homer1 engulfment using Imaris 3D surface rendering shown as: Homer1 volume in CD68+ lysosomes in P2Y12+ microglial surface/P2Y12 volume x100. Transparent points = individual microglia (5–6 microglia per mouse; 24 WT control, 24 NL-F KI control, 24 WT test and 34 NL-F KI test injected microglia were sampled in total), full points = mouse average of ROIs (n=4–6 male mice per condition). Two-way ANOVA (Interaction: F (1, 14) = 4.929, p<0.05) followed by Bonferroni’s multiple comparisons post-hoc test on Y=log(Y) transformed mouse average. I) Representative images of excitatory post-synaptic Homer1 (green) and pre-synaptic Bassoon (magenta) immunostaining in the 6-mo WT and NL-F KI CA1 SR hippocampus post control or test CRISPR-saCas9 injection using super-resolution Airyscan confocal microscopy. Scale bar = 1 μm. Insets show regions of colocalization. J) Quantification of the number of colocalized Homer1 and Bassoon spots shown as density per μm3 using Imaris. Transparent points = individual ROIs (3 ROIs per mouse; 12 WT control, 12 NL-F KI control, 12 WT test and 15 NL-F KI test injected ROIs were sampled in total), full points = mouse average of ROIs (n=4–5 male mice per condition). Two-way ANOVA (Interaction: F (1, 13) = 11.02, p<0.01) followed by Bonferroni’s multiple comparisons post-hoc test on Y=log(Y) transformed mouse average. All data shown as mean ± SEM. p-values shown ns P>0.05; *P<0.05; **P<0.01; ***P<0.001; ****P<0.0001.