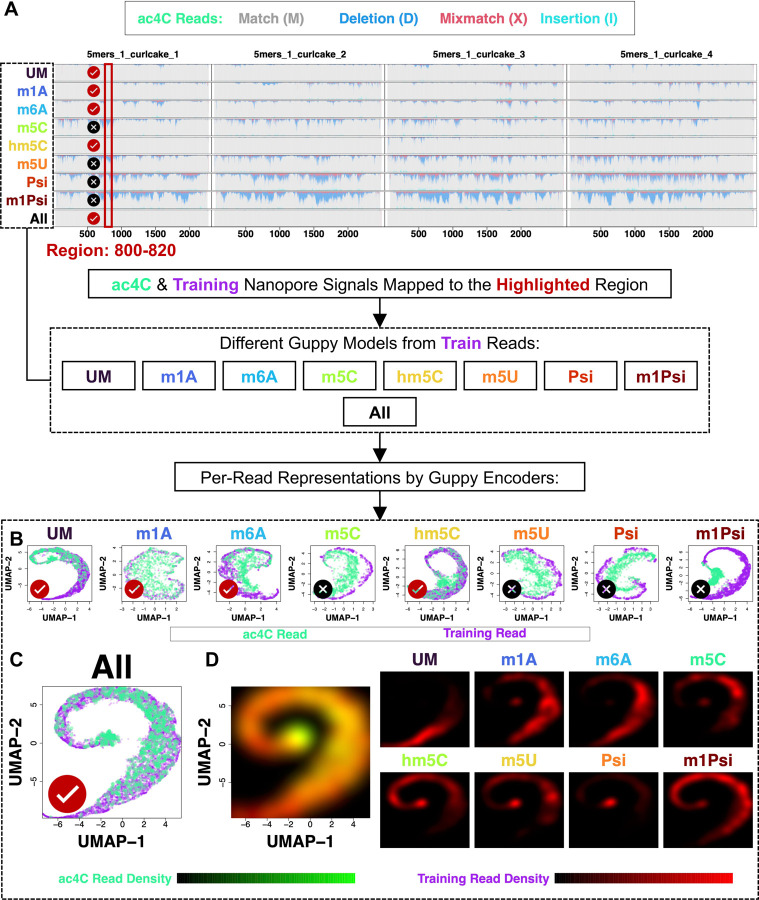
Figure 4. Diverse training data expands the representation space thus making the basecaller generalizable to novel modifications.
(A) Performance of individually and jointly-trained basecallers on ac4C reads was visualized with the genome viewer graph, which shows per-nucleotide CIGAR fractions. All, the jointly-trained basecaller by all the oligo types except for ac4C; other acronyms denote individually-trained basecallers. For individually (B) and jointly-trained (C) basecallers, read fragments mapped to the boxed region were first converted as representation vectors with different basecaller encoders, then visualized by a UMAP plot. Train denotes reads used for training the corresponding basecaller. (D) Spatial distributions of different oligo types in the UMAP space as shown in (C). Black-to-green and red palette denotes ac4C and training reads, respectively.