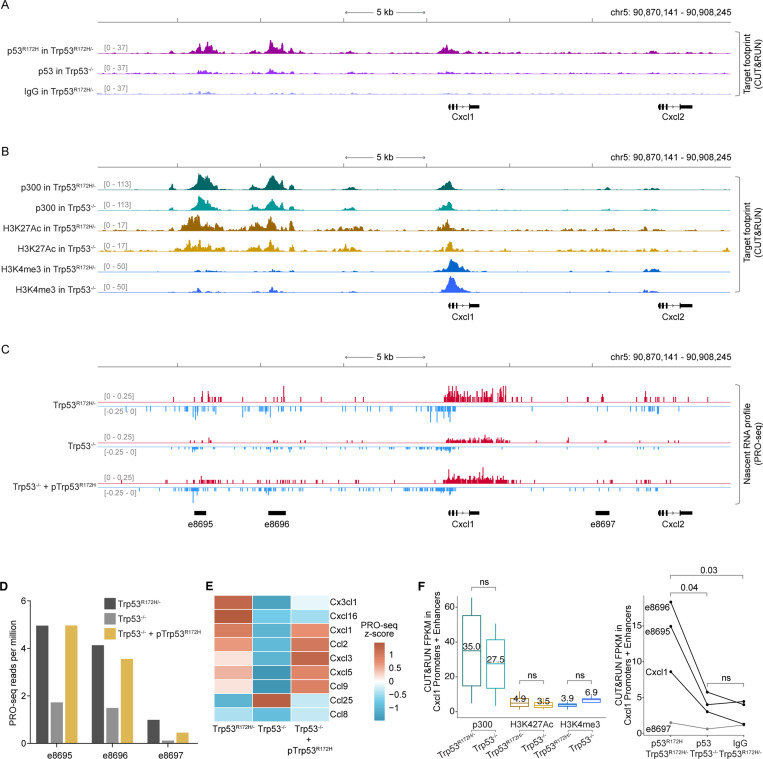
Figure 4. p53R172H occupies and modulates Cxcl1 enhancers.
A, p53R172H occupancy at and around the Cxcl1 gene in Trp53R172H/− and Trp53−/− cells using CUT&RUN. Non-specific IgG in Trp53R172H/− cells is used as a control.
B, p300, H3K27Ac, and H3K4me3 occupancy at and around the Cxcl1 gene in Trp53R172H/− and Trp53−/− cells using CUT&RUN.
C, Nascent RNA profiles at and around the Cxcl1 gene in Trp53R172H/−, Trp53−/−, or Trp53−/− + pTrp53R172H cells using PRO-seq. Nascent RNA profiles are used to call de novo enhancers using dREG71, and the annotated enhancers are shown at the bottom (names begin with “e”).
D, Quantification of enhancer RNA (eRNA) from PRO-seq in the three enhancers around the Cxcl1 gene.
E, Nascent RNA levels of the expressed Chemokine genes shown as the z-cores heatmap of PRO-seq RPM in the three isogenic cells.
F, Quantification of p300 and histone modification levels (left) and p53R172H occupancy (right) at the promoter and enhancers of the Cxcl1 gene using CUT&RUN. P-values are calculated excluding e8697 using a paired t-test.