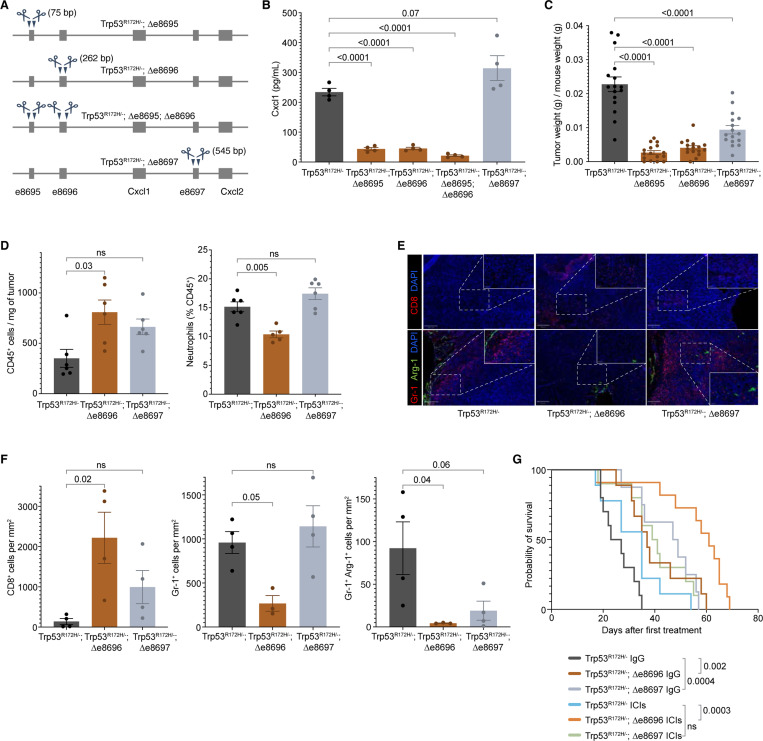
Figure 5. p53R172H-occupied Enhancers regulate Cxcl1 expression and dictate Cxcl1-mediated immunosuppression.
A, Generation of enhancer-deleted isogenic cells from the parental Trp53R172H/− cells. Two enhancers upstream of the Cxcl1 gene and one downstream of the Cxcl1 gene are deleted individually or in a pair using CRISPR/Cas9. The size of deleted regions is indicated in parentheses.
B, Quantification of the Cxcl1 chemokine level in enhancer-deleted isogenic cells and comparison with the Trp53R172H/− cells. P-values are calculated from a one-way ANOVA test followed by a post hoc test with Benjamini-Hochberg correction.
C, Weights of the enhancer-deleted isogenic tumors compared with the Trp53R172H/− tumors. Trp53R172H/−;Δe8695;Δe8696 cells were not implanted in mice due to the minimal effect over either Trp53R172H/−;Δe8695 or Trp53R172H/−;Δe8696 cells. P-values are calculated from a one-way ANOVA test followed by a post hoc test with Benjamini-Hochberg correction.
D, Effect of the e8696 and e8697 status in leukocyte and neutrophil infiltration in PDAC tumors. P-values are calculated from a one-way ANOVA test followed by a post hoc test with Benjamini-Hochberg correction.
E, Representative multiplex immunofluorescence staining of Trp53R172H/−;Δe8695 and Trp53R172H/−;Δe8696 tumors. The scale bars correspond to 200 μm and 50 μm in the main view and the magnified view, respectively.
F, Quantification of immune cell density from randomly selected 500 × 500 μm regions. P-values are calculated from a one-way ANOVA test followed by a post hoc test with Benjamini-Hochberg correction.
G, Kaplan-Meier survival curves showing the effects of e8696 or e8697 status and ICIs in the survival of mice implanted with parental Trp53R172H/− or Trp53R172H/−;Δe8696, or Trp53R172H/−;Δe8697 cells. Control mice were treated with IgG. Trp53R172H/− mice survival cohort is the same as in Figure S4D & Figure 3I. P-values are calculated using a log-rank (Mantel-Cox) test.