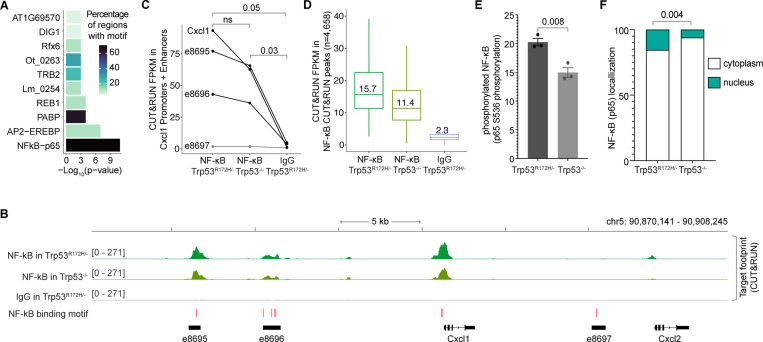
Figure 6. p53R172H occupancy in Cxcl1 Enhancers is dependent on NF-κB.
A, Enriched transcription factor motifs at the promoters and enhancers of the p53R172H-dependent chemokine genes identified using HOMER105.
B, NF-κB occupancy at the Cxcl1 gene and enhancers in Trp53R172H/− and Trp53−/− cells using CUT&RUN. Non-specific IgG is used as a control. The HOMER-identified NF-κB motifs are shown at the bottom (red bars).
C, Quantification of NF-κB occupancy at the promoter and enhancers of the Cxcl1 gene using CUT&RUN. P-values are calculated excluding e8697 using a paired t-test.
D, Quantification of NF-κB occupancy at the NF-κB CUT&RUN peaks using CUT&RUN.
E, Quantification of NF-κB phosphorylation in the Trp53R172H/− and Trp53−/− cells using ELISA on cell extracts. The antibody targets S536 in the p65 subunit of NF-κB.
F, Quantification of NF-κB localization in the cytoplasm vs nucleus using western blot (serial dilution of protein lysates). Vinculin and histone H3 were used as the markers of cytoplasmic and nuclear fractions, respectively. P-values are calculated from a two-way ANOVA test.