Figure 2.
Unsupervised annotations of the DKD Kidney samples
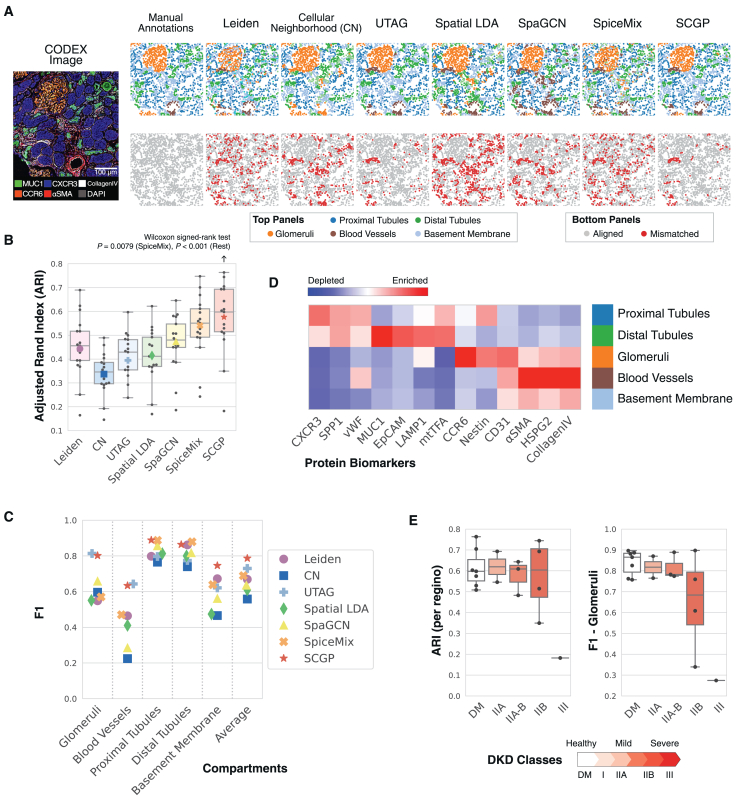
(A) Annotations from SCGP and other unsupervised annotation methods recognized tissue structures aligned with manually annotated compartments. Nodes representing cells are colored according to the assigned clusters/partitions in the top images, and colors not listed in the legend (e.g., cyan) refer to clusters/partitions that cannot be matched to any compartment. Mismatched nodes are highlighted in red in the bottom images.
(B) ARIs were calculated between unsupervised annotations and manual annotations. SCGP performed significantly better than all other methods (Wilcoxon signed-rank test).
(C) For each manually annotated compartment, F1 scores were calculated between manual annotations and the most overlapped cluster/partition.
(D) Signature protein biomarkers for SCGP-identified partitions match expectations of kidney tissue structures.
(E) SCGP annotations on samples with different classes of DKD show varying levels of alignment and accuracy. Diabetes mellitus (DM, with healthy kidney) and DKD classes represent the different progression stages assigned by a nephrologist following the Tervaert classification.41
See also Figure S2.