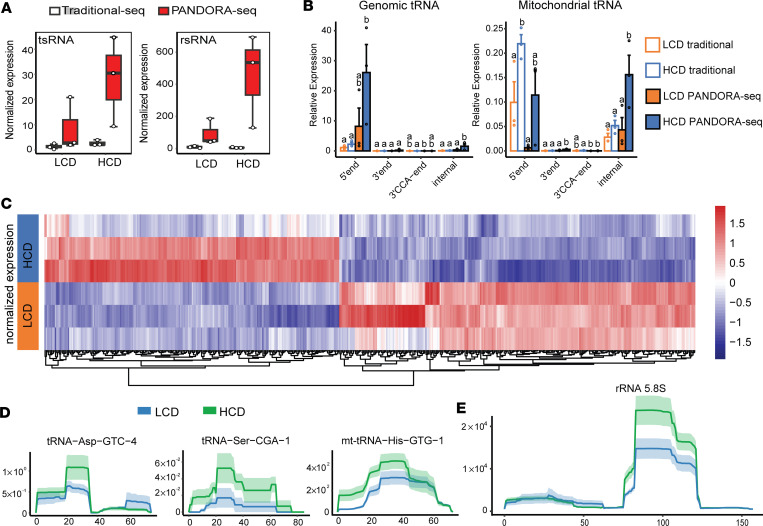
Figure 9. PANDORA-Seq reveals significantly changed sperm tsRNAs and rsRNAs induced by high-cholesterol diet feeding in male LDL receptor–deficient mice.
Three-week-old male LDLR–/– mice were fed an LCD or HCD for 9 weeks. Total RNAs were isolated from the sperm and used for PANDORA-Seq and traditional small RNA sequencing. (A) Sperm tsRNA and rsRNA relative expression (normalized to miRNAs) under traditional sequencing and PANDORA-Seq protocols. (B) Sperm tsRNA responses to traditional sequencing and PANDORA-Seq in regard to different genomic or mitochondria tRNA origins (5′tsRNA, 3′tsRNA, 3′tsRNA-CCA end, and internal tsRNAs). The y axes represent the relative expression levels compared with total reads of miRNA. Different letters above the bars indicate statistically significant differences (P < 0.05). Same letters indicate P > 0.05. Statistical significance was determined by 2-sided 1-way ANOVA with uncorrected Fisher’s least significant difference test. All data are plotted as mean ± SEM. (C) Heatmap representation of differentially expressed sperm tsRNAs detected by PANDORA-Seq. Biological replicates are represented in each row. Red represents relatively increased expression, whereas blue represents decreased expression with adjusted P < 0.05 and FC > 2 as the cutoff threshold. (D and E) Dynamic responses to LCD or HCD of representative sperm tsRNAs (D) and rsRNAs (E) detected by PADNORA-Seq. Mapping plots are presented as mean ± SEM (n = 3 in each group).