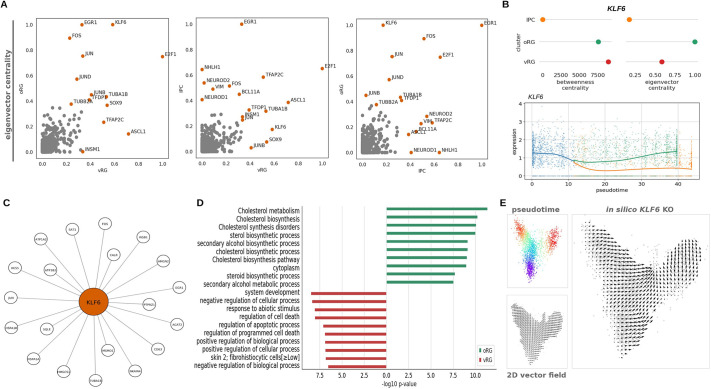
Fig. 3.
Gene regulatory network reconstruction from human neural progenitor single-cell data. (A) Pairwise comparisons of eigenvector centrality values among single-cell progenitor cell clusters, highlighting top 10 genes in each cluster. Some differentially expressed genes for the ventricular radial glia (vRG) cell cluster retain some level of expression in basal progenitors and are indeed present among top ten genes for different gene regulatory network (GRN) connectivity measures across clusters (see also Fig. S7); this is the case for EGR1, FOS or JUN. In addition to KLF6 for outer radial glial cells (oRG), other genes that are more prominently associated with specific clusters include ASCL1, SOX9, TFAP2C for vRG when compared with oRG, or neuron differentiation-related basic helix-loop-helix transcription factors NEUROD1, NEUROD2 and NHLH1 between intermediate progenitor cell (IPC) and radial glia clusters, consistent with the closer transcriptomic similarity of IPCs to excitatory neurons (Bhaduri et al., 2021). (B) KLF6 network measures across single-cell clusters, with a marked contrast between IPC and RG clusters, and most prominently as central node in oRG (eigenvector centrality). Below, KLF6 expression along pseudotime, showing upregulation in oRG and downregulation in IPC. (C) Top representative genes by network weight among KLF6 target genes. (D) Top GO terms associated with KLF6 targets in oRG and vRG, with prominence of cholesterol metabolism in oRG. Cholesterol metabolism GO terms only appear for vRG cluster KLF6 targets if relaxing the P-value threshold above 0.01 (see also Table S3). (E) A vector field represents the predicted bifurcation trajectory from apical (low pseudotime values) to basal (high pseudotime values) progenitors. The in silico perturbation of KLF6 predicts a depletion of both oRG and vRG, with a cell fate shift towards IPC.