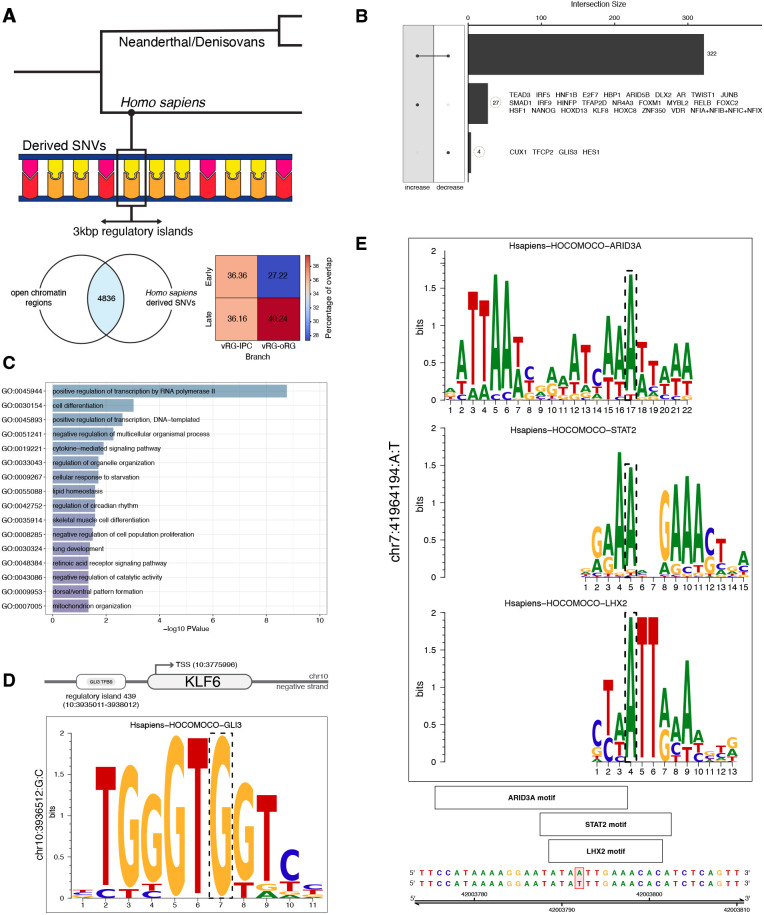
Fig. 4.
Paleogenomic analysis of regulatory variants. (A) Building on the brain atlas of open chromatin regions, regulatory islands were defined as 3 kbp-length regions where Homo sapiens acquired derived alleles in comparison to Neanderthals and Denisovans (carrying the ancestral version found in chimpanzees). Genes associated with regulatory islands are found in pseudotime-informed non-negative matrix factorization (piNMF) modules detected for both ventricular radial glia (vRG) to intermediate progenitor cell (IPC) and vRG to outer radial glia (oRG) branches, with more pronounced abundance on the oRG lineage. (B) Overview of the changes in transcription factor (TF) binding affinity scores to target sites harboring Homo sapiens-derived regulatory variants. (C) GO enrichment analysis for TFs whose binding affinity is impacted by Homo sapiens-derived regulatory variants. (D) Schematic visualization of the predicted TF differential regulation of KLF6 by GLI3, with TF motif impacted by a Homo sapiens-derived single nucleotide variant (dashed box). (E) Schematic visualization of TF binding sites with Homo sapiens-derived mutations highlighted in the context of GLI3 (dashed box).