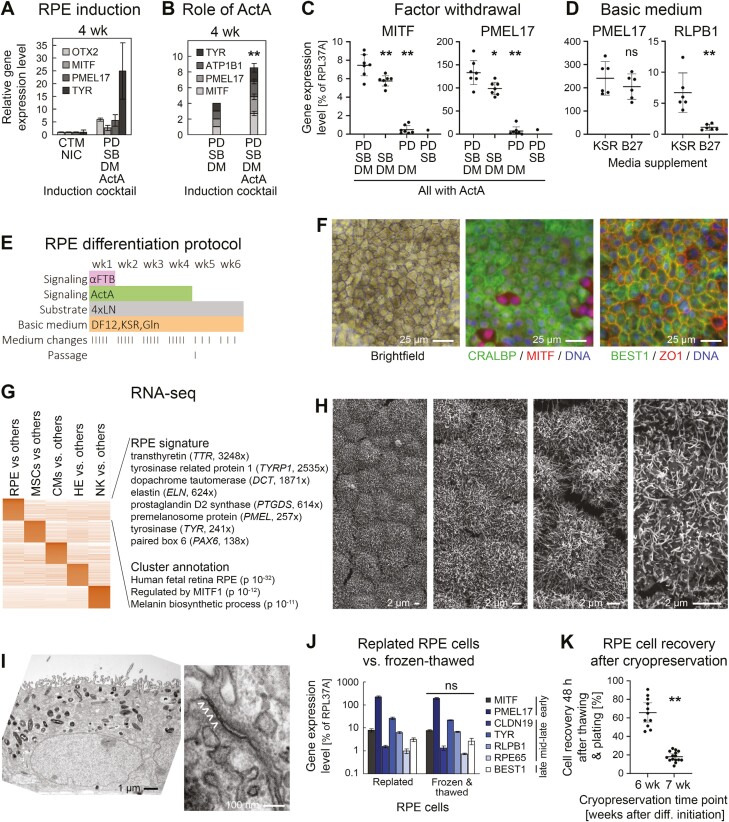
Figure 2.
Directed all-2D RPE induction. (A) RT-qPCR analysis following RPE induction with 2 protocols (line R26, n = 2-6 per data point). PD: PD0325901 (αFGF), SB: SB431542 (αTGFβ), DM: Dorsomorphin (αBMP). (B) RT-qPCR data showing importance of Activin A supplementation in optimized induction cocktail (line R26, n = 5 for + ActA). (C) Analysis of individual contributions by 3 small molecules in the RPE induction cocktail (RT-qPCR data at 4 weeks). (D) Comparison of media supplements used in the new differentiation protocol. Low RLPB1 expression was consistently observed with the B27 supplement. (E) Optimized RPE differentiation protocol. The FGF, TGFβ, and BMP pathways were (moderately) inhibited during the first week. Overlapping ActA treatment was productive for RPE induction. (F) Light and immunofluorescence analysis of maturated iPSC-RPE cells. A minority of (likely more immature) cells was MITFhigh/CRALBP-negative. (G) RNA-seq analysis of iPSC-RPE cells against other derivatives. Selected markers with ratios against other cell types as well as annotation terms of the RPE cluster are highlighted. (H) Scanning EM pictures at different magnifications reveal pronounced microvilli on all RPE cells. (I) Transmission EM analysis reveals apical-basal polarity with biased localization of melanosomes and nuclei (left) as well as tight junctions (right, marked with arrow heads). (J) Freeze-thawing preserves RPE marker expression (RT-qPCR analysis, n = 2). K, iPSC-RPE cells can efficiently be recovered from cryopreserved stocks by 6 but not 7 weeks of differentiation. All data in this figure are based on iPSC line R26.