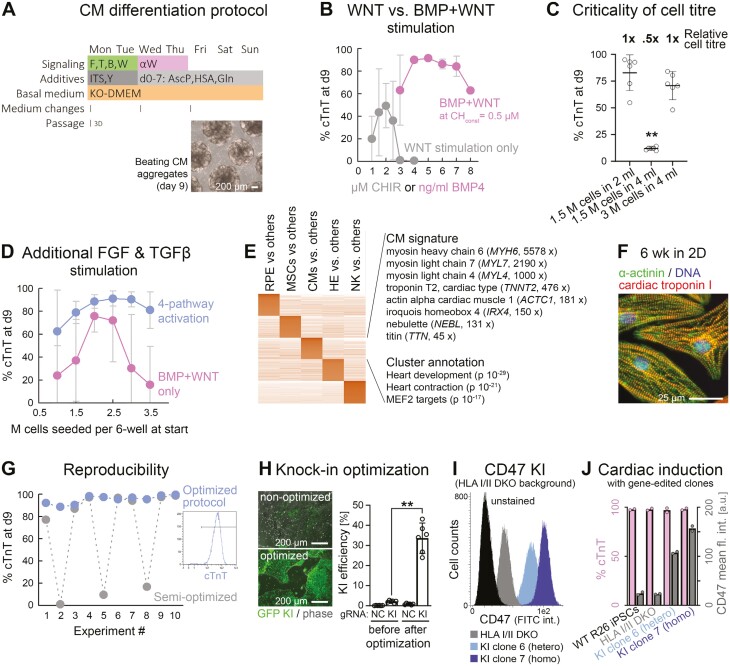
Figure 4.
High-efficiency cardiac induction hallmarked by improved robustness. (A) Outline of optimized protocol based on initial co-stimulation of the FGF, TGFβ, BMP, and WNT pathways. Differentiation commenced upon—not after—EB formation. (B) BMP and WNT co-stimulation (without FGF/ TGFβ) enables superior CM differentiation as compared to WNT stimulation alone, in this protocol (n = 2-10 per data point). (C) Critical dependency of BMP + WNT protocol on cell titer (flow cytometry data). (D) Additional FGF and TGFβ pathway stimulation enhances protocol robustness with regard to cell titer dependency (n = 6-8—dropouts were scored as 0%). (E) Markers and annotation terms of cardiac differentiation cluster (RNA-seq analysis). (F) Immunostaining of iPSC-CMs maturated under adherent conditions. (G) Fully optimized procedure overcomes inter-experimental variation. Data shows flow cytometry data of 10 independent experiments conducted in a row. (H) Optimization of CRISPR-mediated knockin efficiencies in iPSCs using a GFP vector targeting the AAVS1 locus. NC: Negative control using an irrelevant gRNA. (I) Representative flow cytometry plot showing negative controls as well as isolated hetero and homozygous knockin clones of a CD47 transgene targeted to AAVS1 on an HLA I/II deficient background. Note the correlation between signal and gene dosage. (J) Analysis of these iPSC lines after directed cardiac differentiation according to panel A. Data imply consistent differentiation after KO/KI editing and preserved gene dosage-dependent transgene expression.