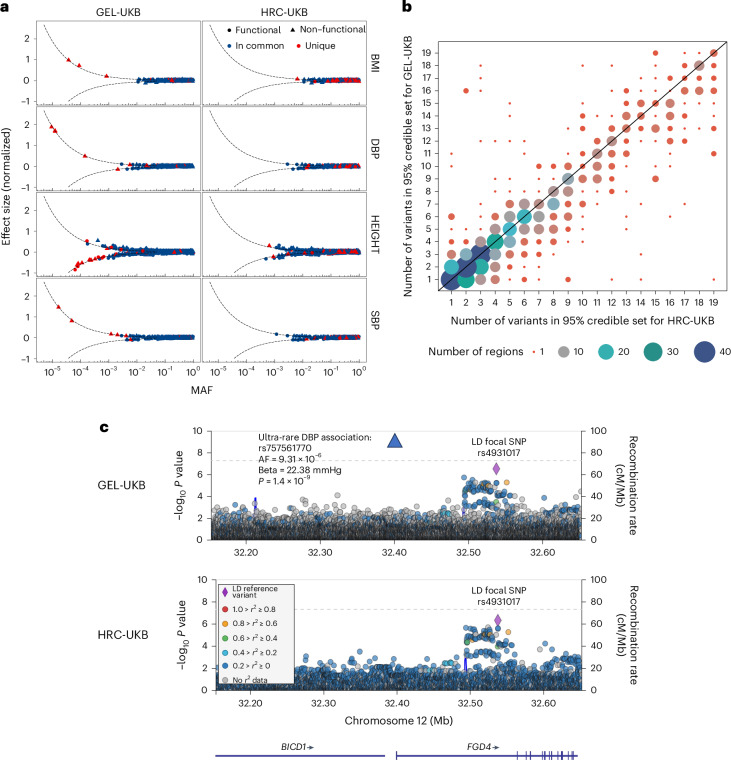
Fig. 2. GEL-imputed UK Biobank data boost power to find common and rare associations.
a, A set of independent genome-wide significant (P < 5 × 10−8) associations identified by step-wise regressions (conditioned joint analysis), and with INFO > 0.8, are plotted versus their imputed allele frequency (x axis). Blue points represent variants that were flagged by step-wise regressions in one dataset and also showed a significant GWAS association in the other dataset. Red points indicate variants unique to that dataset. The shape of the data points reflects the predicted consequences of the variants as determined by the Ensembl Variant Effect Predictor (release 105)14. Dots represent functional variants, including stop gained, stop lost, splice donor/acceptor, frameshift, in-frame insertion/deletion and missense variants and the triangles indicate non-functional variants. The dashed lines indicate the smallest hypothetical effect sizes that can be captured by the P-value threshold (P < 5 × 10−8) at power of 0.5. b, Comparison of the number of variants in the 95% credible sets for GEL-UKB and HRC-UKB fine-mapping results for standing height (capped at 20 variants; Methods). The circle sizes represent the number of fine-mapping regions showing each combination; plots below the diagonal correspond to GEL-UKB having fewer variants in the credible set compared to HRC-UKB. c, LocusZoom plot of ultra-rare-variant association (rs757561770) (in blue triangle) detected by GEL-UKB. The color indicates the linkage disequilibrium (LD) between SNPs and the focal SNP rs4931017, showing that rs757561770 is in low linkage disequilibrium with the focal SNP (r2 = 6.57 × 10−6). Blue lines show the regional recombination rate.