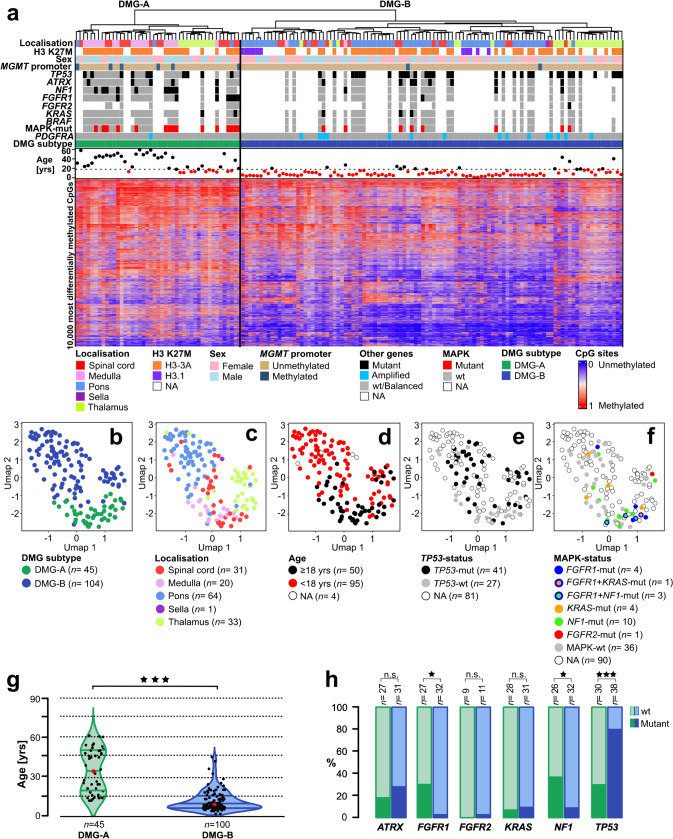
Fig. 1.
DMGs epigenetically split into two subtypes DMG-A and DMG-B that differ with respect to age, tumour localisation, TP53-mutations and MAPK-signalling pathway alterations. a Unsupervised hierarchical clustering of global DNA methylation data from 149 H3 K27M-mutant DMGs of four different localisations (spinal cord n = 31, medulla n = 20, pons n = 64, thalamus n = 33; one patient with an additional sellar tumour). DMGs subdivided into two clusters, corresponding to two DMG-subtypes DMG-A and DMG-B, that were enriched for different features. DMG-A (green, n = 45) was enriched for a medullary localisation (40.0%; n = 18 medullary cases), MAPK-associated mutations (55.6%; n = 15/27 cases sequenced) and cases with a methylated MGMT promoter (13.3%; n = 6/45). DMG-B (blue, n = 104) contained many pontine tumours (58.7%; n = 61) and TP53-mutant cases (78.9%; n = 30/38 cases sequenced). Most of the FGFR1-mutant cases formed a subcluster in the DMG-A cluster. b–f Uniform Manifold Approximation and Projection (UMAP) of the same cases shows similar results. b Again, DMG-A (green) and DMG-B (blue; subtype attribution from a) separated, as well as medullary versus pontine cases (c) and adult versus paediatric patients (d). e TP53-mutant cases were enriched in the part of the UMAP containing the DMG-B cases. f Most FGFR1- and NF1-mutant DMG were found in close proximity, mixed with few DMGs without known MAPK-associated alteration. g Violin plot of patient age. DMG-A (left) showed a bimodal age distribution with a median age at diagnosis of 31.0 ± 16.0 years. Patients with DMG-B were significantly younger (right; median age 7.6 ± 7.6 years; p < 0.001). The lines within the violin plots represent the quantiles (0.25, 0.50, 0.75), the red dots the median. h Distinct mutations were enriched in the two DMG-subtypes: TP53-mutations were enriched in DMG-B, while mutations associated with the MAPK-signalling pathway were enriched in DMG-A. Significance levels: *p ≤ 0.05, **p ≤ 0.01, ***p ≤ 0.001