Figure 4.
IgG binding profiles in N.LT and S.LT rabbits
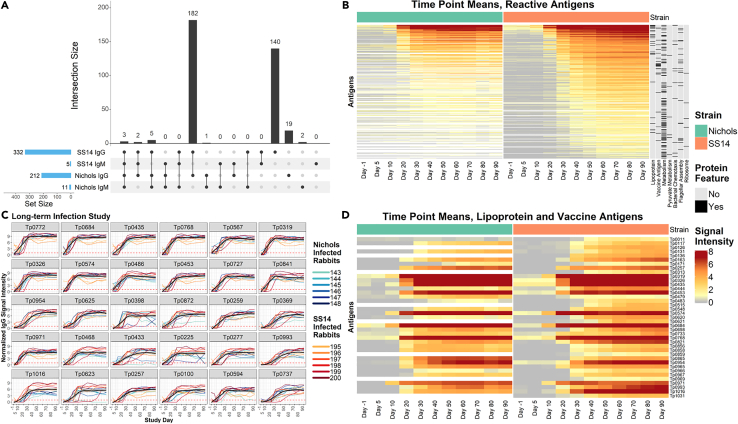
(A) UpSet plot summarizing both the set and set size of antigens recognized by IgG and IgM during experimental infection of rabbits with either the Nichols or SS14 strains of T. pallidum. The UpSet plot is equivalent to a Venn diagram, where each vertical bar represents antibody responses that “intersect” or overlap between two or more categories (i.e., antigens that are reactive in each of the categories specified by the connected dot matrix below the bar graph). The blue horizontal bars represent the total number of reactive antigens in each category.
(B) Heatmap showing the mean IgG binding signal intensity for each reactive antigen in either Nichols or SS14-strain-infected rabbits. Columns represent the means of each group of rabbits at each sequential time point, and rows represent T. pallidum proteins sorted by hierarchical clustering. Protein features that were significantly associated with antibody reactivity in stepwise regression models are shown in the black and gray columns to the right of the heatmap. Responses are grouped by Nichols- (green header) and SS14 (orange header)-strain-infected rabbits.
(C) Line plots of reactivity to the 30 most reactive antigens in animals infected long-term with the Nichols strain (teal to dark blue lines) and the SS14 strain (orange to dark red lines). In each graph, the black line represents the running mean of all samples at each time point post-infection.
(D) Heatmap of the antigens annotated as lipoproteins or OMPs/vaccine candidates (ordered by gene ID) recognized in Nichols-infected animals (left side, green header), and SS14-infected rabbits (right side, orange header) over the 90-day infection period. Data are represented as mean ± SEM.