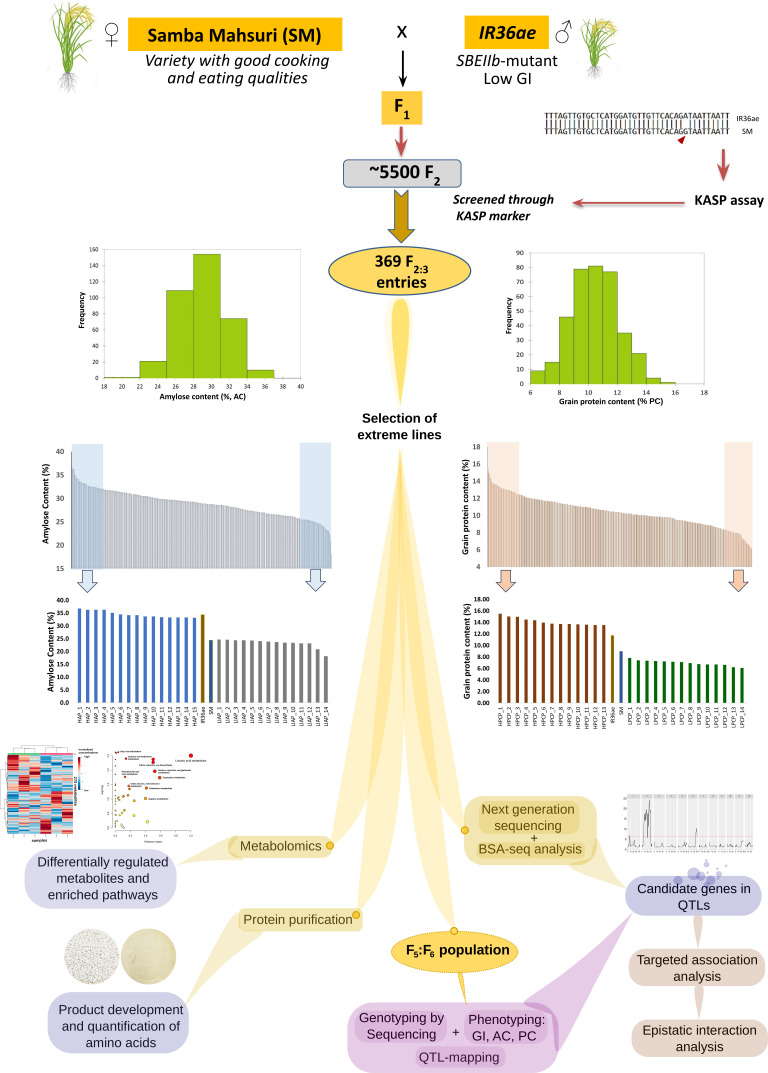
Fig. 1.
Analytical workflow of RIL population development and downstream multigenomic analysis pipeline for identifying candidate genes and genotypes with ultralow and low GI, high AC, and high PC. The RIL population resulting from the cross between Samba Mahsuri and IR36ae carrying mutation for starch branching enzyme IIb (sbe IIb) gene was initially screened using kompetitive allele-specific PCR (KASP) markers among F2:F3 lines. Highly contrasting lines with extremely high and low AC and PC were selected for further analysis using next-generation sequencing for bulk segregant QTL (BSA-seq) analysis. RIL population advancement made until F6 generation subjected to genotyping-by-sequencing method, phenotyping, and QTL mapping in order to determine candidate genes and important lines possessing ultralow/low GI, high or intermediate AC, and high PC which were then subjected to metabolomics analysis and protein purification for further product development in diabetes-stricken communities, especially in low- and middle-income regions.