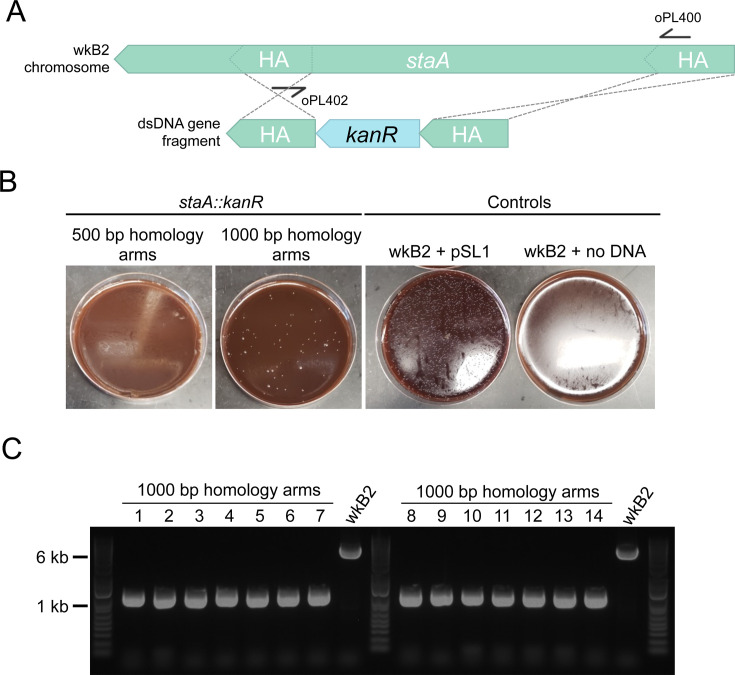
Fig 2.
Gene knockout in staA is accurate. (A) Gene deletion map depicting genomic staA and the corresponding deletion construct. The deletion construct is composed of a kanR cassette flanked by 500 or 1,000 bp arms with homology internal to the staA gene. Primers oPL400 and oPL402 were designed for use in PCR screening for proper deletion. (B) Images of staA::kanR transformants and controls on Columbia + 5% sheep’s blood (Col-B) + kanamycin agar plates. wkB2 can be successfully transformed with a staA::kanR cassette containing 1,000 bp (middle left), but not 500 bp (left), homology arms, indicating that 1,000 bp of homology is needed to isolate colonies with the deletion. wkB2 cannot grow on kanamycin plates (right) without the presence of a kanamycin resistance cassette, as illustrated by control cells that received the kanR-containing pSL1 plasmid (middle right). (C) DNA gel of PCR screen for staA deletion. Large and small colonies of wkB2 + staA::kanR with 1,000 bp homology arms or wkB2 were amplified via PCR using oPL400 and oPL402, and run on a 1% agarose gel. The ~6,000 bp band observed in the wkB2 sample corresponds to the genomic region within staA, whereas the ~1,000 bp band observed in experimental samples corresponds to the kanR cassette. 100% of screened colonies were found to contain a single 1,000 bp band, indicating proper deletion of staA.