FIGURE 1.
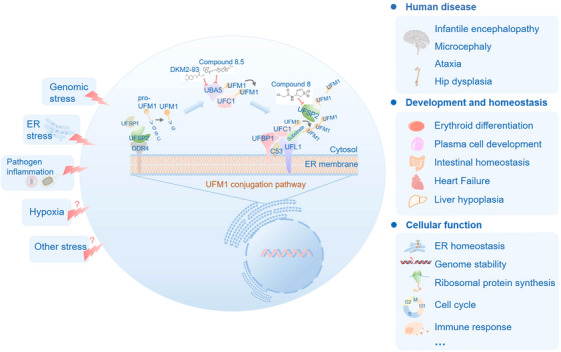
The regulatory system of UFMylation. Pro‐UFM1 is cleaved by UFSP1 and UFSP2 in its C‐terminal region, which generates the mature UFM1 by exposing the C terminus glycine residue. UBA5 activates UFM1, and transfers to UFC1, the E2 conjugating enzyme. Subsequently, the UFM1‐UFC1 thioester intermediate is transferred to a stable UFM1‐specific E3 enzyme complex comprising UFL1 and UFBP1. This complex is anchored on the cytosolic side of the ER. CDK5RAP3 (C53 in the Figure) acts as a stabiliser for the complex at the ER. Both UFBP1 and CDK5RAP3 are possible adaptor proteins that allow the ligase UFL1 to recruit a wider pool of substrates. Eventually, the substrates become mono‐ or poly‐UFMylated. The UFMylated substrates are cleaved mainly by UFSP2, which is a major de‐UFMylation enzyme of the pathway, and forms a complex with ODR4 to localise to the ER. Consistent with conservation of the UFM1 system, genetic screenings across diverse human populations have identified variants of the UFM1 system genes in neurodevelopmental diseases, including infantile encephalopathy, microcephaly and ataxia, 39 , 40 , 41 hip dysplasia 42 , 43 and carcinomas. 104 Genetic deletion of key UFM1 system components, such as UBA5, UFL1, UFBP1 or CDK5RAP3, revealed that the UFM1 system is critical to mice embryonic development, plasma cell development, haematopoiesis, erythroid differentiation, liver development, intestinal homeostasis, heart failure and zebrafish epiboly. 12 , 16 , 29 , 31 , 44 , 45 , 46 , 47 Importantly, the identification of numerous UFMylation substrates underscores the critical role of UFMylation in multiple cellular functions, such as ER homeostasis, including ER‐RQC and ER‐phagy, genomic stability, tumorigenesis and immune response.
