Fig. 4. Geometric cues organize the alignment of contractile force along the FA-nuclear axis to modulate nuclear stiffness and YAP nuclear localization.
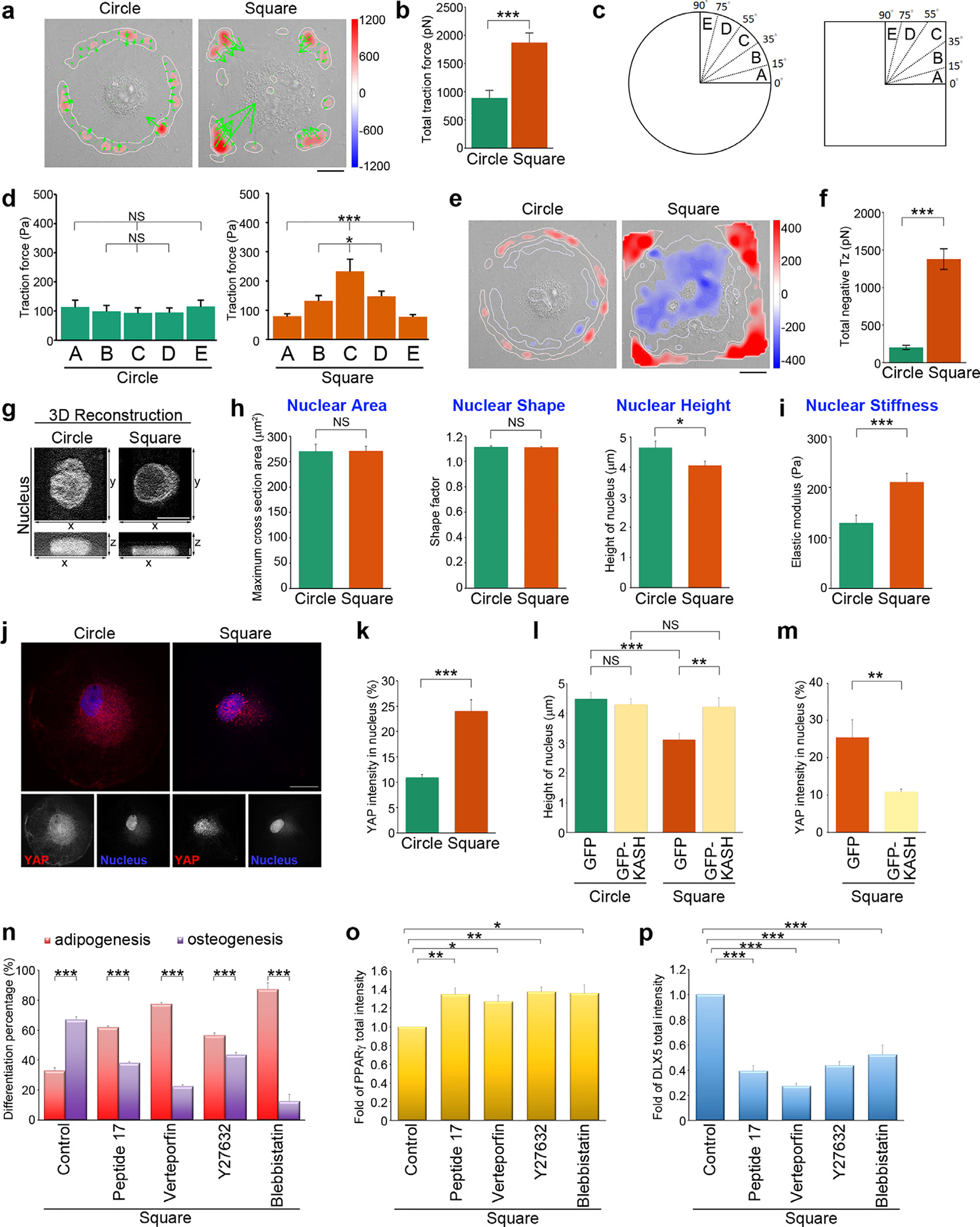
(a) Image of the traction force distribution of MSCs plated on circular or square micropatterns after cultured for 16 hrs in growth medium. Scale bar, 20 μm. (b) Total traction force (pN) of MSCs, as indicated in (a). Data are mean ± s.e.m [n = 10 cells (circle) and 10 cells (square) from 3 independent experiments]. Data was tested for normality and homoscedasticity in SPSS. Statistical analysis of data was carried out by the Student’s t-test. ***p < 0.001. (c) A schematic diagram of angular sectors used for segmenting an imaged cell. (d) Tangential traction force (Pa) at 0–15 μm from edge of MSCs, as indicated in (a), within the angular sectors of the first quadrant. Data are mean ± s.e.m [n = 10 cells (circle) and 10 cells (square) from 3 independent experiments]. Data were tested for normality and homoscedasticity in SPSS. Statistical analysis of data was carried out by one-way ANOVA, followed by the Student’s t-test. *p < 0.05; ***p < 0.001; NS, no significance. (e) Maps of out-of-plane traction stresses of MSCs plated on circular or square micropatterns after cultured for 16 hrs in growth medium. Scale bar, 20 μm. (f) Total compressive stresses (pN) of MSCs, as indicated in (e). Data are mean ± s.e.m [n = 10 cells (circle) and 10 cells (square) from 3 independent experiments]. Data were tested for normality and homoscedasticity in SPSS. Statistical analysis of data was carried out by the Student’s t-test. ***p < 0.001. (g) 3D reconstruction taken from the z-series of confocal images of MSCs stained with DAPI (to visualize nucleus) at every 0.2 μm from top to bottom. The top view (x-y) and side view (x-z) images of the nuclei in MSCs plated on circular or square micropatterns after cultured for 16 hrs in growth medium. The top view: scale bar, 20 μm; the side view: scale bar, 2 μm. (h) Left: Projected area of the nuclei from MSCs, as indicated in (g). Data are mean ± s.e.m [n = 29 cells (circle) and 29 cells (square) from 3 independent experiments]. Data were tested for normality and homoscedasticity in SPSS. Statistical analysis of data was carried out by the Student’s t-test. NS, no significance. Middle: Shape factor of nucleus from MSCs, as indicated in (g). Data are mean ± s.e.m [n = 29 cells (circle) and 29 cells (square) from 3 independent experiments]. Data was tested for normality and homoscedasticity in SPSS. Statistical analysis of data was carried out by the Student’s t-test. NS, no significance. Right: Height of the nuclei from MSCs, as indicated in (g). Data are mean ± s.e.m [n = 30 cells (circle) 28 cells (square) from 3 independent experiments]. Data were tested for normality and homoscedasticity in SPSS. Statistical analysis of data was carried out by the Student’s t-test. *p < 0.05. (i) Intranuclear elastic shear modulus (at 10 Hz; Pa) that quantified the nuclear stiffness for MSCs plated on circle or square micropatterns after cultured for 16 hrs in growth medium. Data are mean ± s.e.m [n = 30 beads/ 24 cells (circle) and 28 beads/26 cells (square) from 3 independent experiments]. Data were tested for normality and homoscedasticity in SPSS. Statistical analysis of data was carried out by the Student’s t-test. ***p < 0.001. (j) Confocal images of MSCs plated on circular or square micropatterns after cultured for 16 hrs in growth medium; there were immunostained with YAP (red) and DAPI (to visualize nucleus; blue). Scale bar, 20 μm. (k) Percentage of YAP average density (intensity per μm2) within nuclei of MSCs, as indicated in (j). Data are mean ± s.e.m [n = 24 cells (circle) and 31 cells (square) from 3 independent experiments]. Data were tested for normality and homoscedasticity in SPSS. Statistical analysis of data was carried out by the Student’s t-test. ***p < 0.001. (l) Height of the nuclei from MSCs overexpressing GFP-C1 (GFP) or the GFP-Nesprin-2 (ΔN)-klarischt sequence (GFP-KASH) and plated on circular or square micropatterns after cultured for 16 hrs in growth medium. Data are mean ± s.e.m [circle, n = 22 cells (GFP) and 25 cells (GFP-KASH); square, n = 21 cells (GFP) and 25 cells (GFP-KASH); the analyzed cells were from 3 independent experiments]. Data were tested for normality and homoscedasticity in SPSS. Statistical analysis of data was carried out by the Student’s t-test. **p < 0.01; ***p < 0.001, NS, no significance. (m) Percentage of YAP average density (intensity per μm2) within the nuclei of MSCs overexpressing GFP-C1 (GFP) or the GFP-Nesprin-2 (ΔN)-klarischt sequence (GFP-KASH) and plated on square micropatterns after cultured for 16 hrs in growth medium. Data are mean ± s.e.m [n = 9 cells (GFP) and 10 cells (GFP-KASH) from 3 independent experiments]. Data were tested for normality and homoscedasticity in SPSS. Statistical analysis of data was carried out by the Student’s t-test. **p < 0.01. (n) Percentage of cells showing osteogenesis (ALP-positive cells) and adipogenesis (Oil Red O-positive cells). MSCs were plated on square micropatterns after cultured for 14 days in mixed differentiation medium containing DMSO (control), peptide 17 (2 μM), Verteporfin (10 μM), blebbistatin (50 μM) or Y27632 (10 μM). Data are mean ± s.e.m (n = 4 independent experiments). Statistical analysis of data was carried out by the Student’s t-test. ***p < 0.001. (o) Fold of PPARγ total fluorescence intensity in the MSCs seeded on square micropatterns after cultured for 7 days in mixed differentiation medium containing DMSO (control), peptide 17 (2 μM), Verteporfin (10 μM), blebbistatin (50 μM) or Y27632 (10 μM). Data are mean ± s.e.m (control, n = 10 cells; peptide 17, n = 19 cells; Verteporfin, n = 26 cells; Y27632, n = 28 cells; blebbistatin, n = 25 cells; the analyzed cells were from 3 independent experiments). Data were tested for normality and homoscedasticity in SPSS. Statistical analysis of data was carried out by the Student’s t-test. *p < 0.05; **p < 0.01. (p) Fold of DLX5 total fluorescence intensity in the MSCs seeded on square micropatterns after cultured for 1 days in mixed differentiation medium containing DMSO (control), peptide 17 (2 μM), Verteporfin (10 μM), blebbistatin (50 μM) or Y27632 (10 μM). Data are mean ± s.e.m (control, n = 20 cells; peptide 17, n = 20 cells; Verteporfin, n = 19 cells; Y27632, n = 20 cells; blebbistatin, n = 19 cells; the analyzed cells were from 3 independent experiments). Data were tested for normality and homoscedasticity in SPSS. Statistical analysis of data was carried out by the Student’s t-test. ***p < 0.001.
