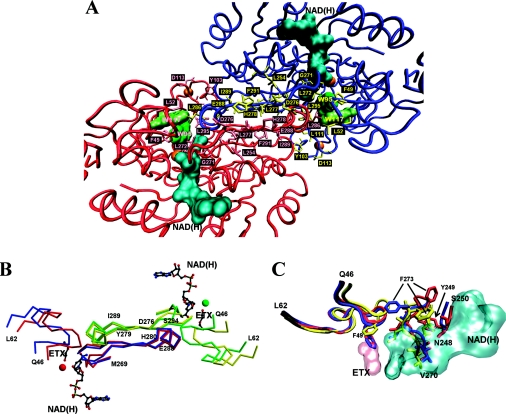
Figure 7. Structural analysis of SsADH and mSsADH.
(A) Partial view of the SsADH holo structure looking at the A–B dimer of the homotetramer, illustrating the helix αF and β-strands, βS and βF of subunit A (red) and B (blue) at dimer interface (centre of Figure). The side chains of the residues involved in the interface interactions are coloured in pink (subunit A) and in yellow (subunit B). The coenzyme molecule and the Trp-95 and Trp-117 side chains are coloured in cyan and green respectively. The orange spheres represent the catalytic and structural zinc atoms. (B) α-Carbon traces of the 46–62 and 269–296 segments from both apo (blue) and holo (red) form of the subunit A, and apo (yellow) and holo (green) form of the subunit B of SsADH, are shown superimposed. Residues 276–279 and 289–294 belong to strand βS and βF respectively, connected by helix αF 280–288. The coenzyme NAD(H) and 2-ethoxyethanol (ETX) molecules are shown in grey. The red and green sphere represents the catalytic zinc in the holo form. (C) Comparison of the regions 46–62, 248–250 and 270–275 of the mutant N249Y SsADH (red) and the apo (blue) and holo (yellow) forms of wild-type SsADH structure. Residues 270–273 interact directly with the NADH molecule (cyan) [10]. In the SsADH apo form, the Phe-49 side chain occupies the same position as the ETX molecule (pink) in the ternary complex structure. The swing of the Phe-273 side chain occurring upon apo–holo transition is shown, as well as its position in the mSsADH apo structure. The movement of the Phe-273 side chain in SsADH allows loop 46–62 to rearrange and extensively interact with the helix αF region of residues 280–286 of the nearby subunit B [10]. Segment 46–62 is one of the most flexible parts of SsADH structure. It was found to be very disordered in the mutant N249Y apo structure due to the conformational changes induced by the mutation [10,11]. Important contacts between this loop and the segment 280–286 of the near subunit B are presumably disturbed or lost in the mutant structure, precluding proper intersubunit communication. This Figure was prepared using VMD [38] and Persistence of Vision Raytracer (http://www.povray.org/) (A, C), MolScript [39] and Raster3D [40] (B).