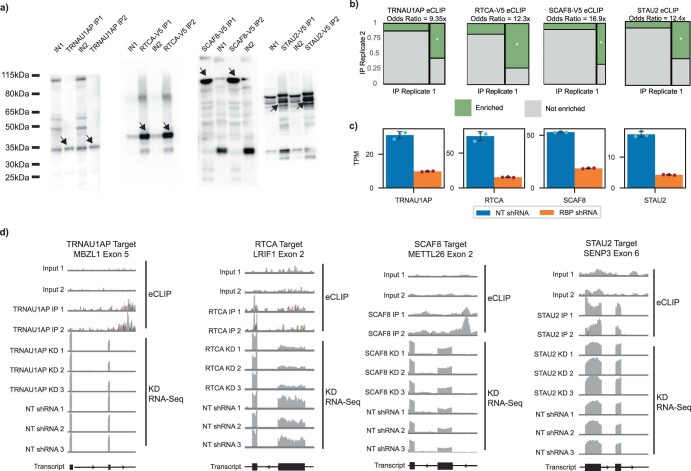
Extended Data Fig. 4. Quality control of eCLIP and shRNA knockdown followed by RNA-seq.
a Western blots of cold gels from eCLIP protocol for TRNAU1AP, SCAF8, STAU2 and RTCA. Size-matched input and immunoprecipitation conditions are compared. n = 2 independent samples, with size-matched input and IP conditions extracted from both. b Mosaic plots from Skipper showing concordance between eCLIP replicates. Odds ratios and significance from Fisher’s exact test. c TPM of unexpected hits following shRNA knockdown as measured from aligned RNA-seq data. (mean ± s.d., n = 3 replicate knockdowns). d IGV browser tracks showing coverage of RBP eCLIP signal relative to sized-matched input and the RBP KD RNA-Seq signal relative to non-targeting shRNA. From left to right: comparison of TRNAU1AP eCLIP and KD RNA-Seq signal near MBZL Exon 5, comparison of RTCA eCLIP and KD RNA-Seq signal near LRIF Exon 2, comparison of SCAF8 eCLIP and KD RNA-Seq signal near METTL26 Exon 2, comparison of STAU2 eCLIP and KD RNA-Seq signal near SENP3 Exon 6.