Abstract
Epstein-Barr virus (EBV) nuclear antigen 3A (EBNA-3A) is essential for virus-mediated immortalization of B lymphocytes in vitro and is believed to regulate transcription of cellular and/or viral genes. One known mechanism of regulation is through its interaction with the cellular transcription factor Jκ. This interaction downregulates transcription mediated by EBNA-2 and Jκ. To identify the amino acids that play a role in this interaction, we have generated mutant EBNA-3A proteins. A mutant EBNA-3A protein in which alanine residues were substituted for amino acids 199, 200, and 202 no longer downregulated transcription. Surprisingly, this mutant protein remained able to coimmunoprecipitate with Jκ. Using a reporter gene assay based on the recruitment of Jκ by various regions spanning EBNA-3A, we have shown that this mutation abolished binding of Jκ to the N-proximal region (amino acids 125 to 222) and that no other region of EBNA-3A alone was sufficient to mediate an association with Jκ. To determine the biological significance of the interaction of EBNA-3A with Jκ, we have studied its conservation in the simian lymphocryptovirus herpesvirus papio (HVP) by cloning HVP-3A, the homolog of EBNA-3A encoded by this virus. This 903-amino-acid protein exhibited 37% identity with its EBV counterpart, mainly within the amino-terminal half. HVP-3A also interacted with Jκ through a region located between amino acids 127 and 223 and also repressed transcription mediated through EBNA-2 and Jκ. The evolutionary conservation of this function, in proteins that have otherwise significantly diverged, argues strongly for an important biological role in virus-mediated immortalization of B lymphocytes.
The ubiquitous human gammaherpesvirus Epstein-Barr virus (EBV) infects epithelial cells in the oropharynx and then establishes a latent infection in B lymphocytes (for a review, see reference 28). These latently infected B cells are believed to constitute the reservoir of virus that sustains the host's lifelong infection. In vitro, EBV-mediated immortalization of B lymphocytes generates lymphoblastoid cell lines in which the full set of viral latent genes is expressed (for reviews, see references 28, 46, and 51). This so-called growth program of latency, or the latency III program, is also observed during the early phase of latent infection in vivo and in EBV-associated lymphoproliferative disorders which occur in immunocompromised patients. The ability to immortalize B lymphocytes in vitro and the association with lymphomas and lymphoproliferative disorders are shared by distantly related lymphocryptoviruses that infect Old World primates such as chimpanzees, baboons, or rhesus macaques (9, 14). Infection of rhesus macaques with their respective virus mirrors EBV infection of humans, making them excellent animal models of EBV infection (39).
Only a very limited number of gene products encoded by EBV can be detected in EBV-immortalized B cells. Three of these are members of the EBV nuclear antigen 3 (EBNA-3) family, two of which, EBNA-3A and -3C, are essential for the establishment of a latent infection in B lymphocytes in vivo (55). An increasing body of evidence indicates that the function of the EBNA-3 proteins, at least in part, is to regulate transcription. The EBNA-3 proteins all bind to the cellular transcription factor Jκ and repress Jκ-mediated transcription by preventing Jκ from binding to DNA (30, 32, 48, 63). Jκ, a highly conserved ubiquitous DNA-binding protein, is the effector component of the Notch signaling pathway, which specifies cell fate during development (for a review, see reference 1). Due to its likely role in development, the molecular mechanism of transcriptional regulation by Jκ is the focus of intense research. Jκ alone does not activate transcription but rather functions as a repressor. A repression domain has been identified in the center of the protein (19), and several potential mechanisms for mediating repression have been proposed. First, by contacting both transcription factor IIA (TFIIA) and TFIID, Jκ is thought to hinder optimal interaction between these two general transcription factors (40). Second, Jκ interacts with a corepressor complex that contains the histone deacetylase HDAC-1 (21, 27, 59). Jκ can also specifically repress NF-κB-mediated transactivation of the NF-κB2 and interleukin-6 genes, by preventing binding of NF-κB to the promoter or its association with a cofactor (26, 41, 43). In addition to its role as a repressor, Jκ can also function as a transcriptional activator. Activation of the Notch receptor, which binds Jκ, releases its intracellular domain complexed to Jκ in a manner that conceals the repression domain and activates transcription. In EBV-transformed cells, EBNA-2 mimics activated Notch by binding to Jκ and activating transcription (15, 20, 35). In Drosophila melanogaster, the homologue of Jκ, Suppressor of Hairless [Su(H)] is regulated by the protein Hairless which prevents binding of Jκ to its consensus sequence (4). The EBNA-3 proteins thus function in a similar manner to Hairless to repress EBNA-2–Jκ-mediated transcription. Regulation by Jκ thus involves multiple mechanisms, mediated through protein interactions. This complexity likely reflects the necessity for tight control of expression of its target genes.
Although many promoters contain the core heptamer recognized by Jκ and represent potential targets, only a limited number have actually been demonstrated to be regulated by Jκ or Su(H). Among these are cellular genes, such as human interleukin-6 (26), murine Hairy Enhancer of Split-1 (22, 23), or the enhancer of split complex genes (2, 33), as well as viral genes such as the adenovirus pIX (8), the EBV LMP-1 and LMP-2 promoters, and the EBV Cp used for transcription of the EBNAs during latency III (17, 24, 31, 56, 60, 61). Jκ is likely a key player in EBV latent infection, especially during latency program III. Not only is Jκ involved in regulation of the expression of most EBV latent genes expressed during latency III but also, because EBNA-2 and the EBNA-3 proteins regulate Jκ-mediated transcription, every gene regulated by Jκ represents a potential target of deregulation by the virus. Clearly, then, regulation of Jκ-mediated transcription is likely to play an important role in the immortalization of B lymphocytes by EBV. One question that remains to be answered is whether all three EBNA-3 proteins are required to regulate Jκ-mediated transcription.
In addition to their biological similarities, the EBV and herpesvirus papio (HVP) genomes exhbit a homologous, colinear organization, and most HVP latent genes have now been cloned (10, 11, 18, 36, 42, 57; B. Zhao, R. Dalbiès-Tran, and C. E. Sample, manuscript in preparation). The global picture emerging from these reports is that, despite a significant overall divergence in the sequence, many elements important for the function of EBV proteins are conserved in their HVP counterparts. Such homology extends to other regions of the genome with essential functions, such as promoters or origins of replication (10, 13, 49). Of particular interest, several features related to the role of Jκ during latent infection are conserved. The EBNA-2 homolog can interact with Jκ and activate transcription from a reporter gene controlled by Jκ binding sites (34, 36). Moreover, the consensus sequence for Jκ is conserved in the simian virus Cp promoter, which, like its EBV counterpart, is responsive to EBNA-2 (13). Whether the homologues of the EBNA-3 proteins would also regulate Jκ-mediated transcription was unknown.
Clearly, the conservation of a given function of an EBV gene product in its HVP homologue is an excellent way to evaluate its biological significance. An alternative way to address this question is to generate a recombinant EBV that is defective in a given function. A recombinant virus encoding a mutant EBNA-3 protein that does not affect Jκ-mediated transcriptional regulation would be a powerful tool for analyzing the biological significance of regulation of Jκ-mediated transcription by EBNA-3. Before we can use either of these techniques, it is necessary to characterize the interaction between EBNA-3 proteins and Jκ and to identify the amino acids that are essential for this interaction. The domain that binds Jκ has been most convincingly mapped for EBNA-3C and is localized to a domain in the amino terminus that is conserved in all EBNA-3 proteins (48, 63). Mutations within conserved amino acids disrupt both binding between EBNA-3C and Jκ, as well as the resultant repression of transcription mediated through Jκ (63). For EBNA-3A, there have been conflicting reports as to whether the corresponding domain binds Jκ. Although we and others have reported that EBNA-3A–Jκ binding is mediated through the conserved domain (3, 63), others have reported binding through a more N-terminal or a more C-terminal domain (6, 48).
Here, we demonstrate that the conserved domain alone was sufficient for interaction with Jκ and that mutations of amino acids conserved between the EBNA-3 proteins prevented this interaction. These mutations also disrupted the ability of EBNA-3A to regulate EBNA-2–Jκ-mediated transcription. Surprisingly, however, the full-length EBNA-3A mutant protein remained able to associate with Jκ, although we were unable to identify a second domain that alone was sufficient to mediate an interaction. Finally, as a measure of the biological relevance of the interaction between EBNA-3A and Jκ, we demonstrate that, although the HVP and EBV 3A proteins share only 37% identity, the interaction of 3A with Jκ was conserved. The conservation of this function suggests that the regulation of EBNA-2–Jκ-mediated transcription by 3A proteins plays a significant role in the life cycle of these viruses.
MATERIALS AND METHODS
Cell lines.
Louckes and BJAB are EBV-negative Burkitt lymphoma and B-lymphoma cell lines, respectively. S594 is an HVP-infected B-cell line derived from the peripheral blood of a baboon. All cell lines were maintained in RPMI 1640 medium supplemented with 2 mM glutamine and 10% fetal bovine serum (HyClone).
Plasmids.
Using the Quickchange site-directed mutagenesis kit (Stratagene) according to the manufacturer's instructions, a HindIII site was introduced into pSG5-Jκ (37) at the end of the coding sequence with the oligodeoxynucleotide primer pair 5′-GCCACAGTGGTATCCAAGCTTCCGTCTTTTTGCTAGGAC and 5′-GTCCTAGCAAAAAGACGGAAGCTTGGATACCACTGTGGC. The HindIII restriction fragment from pCMXVP16 (a gift of Paul Brindle, St. Jude Children's Research Hospital, Memphis, Tenn.), encoding the VP16 activation domain, was cloned into this site.
Mutations in the N-proximal region of EBNA-3A.
Mutations were introduced into pSG5-EBNA3A (63) using the Quickchange mutagenesis kit, with the primer pairs 5′-GCGGGTGCCCAGGCATACAGCAGCTGG and 5′-GGCACCCGCAGGCCATCCGGCGGCCAGGG (resulting in mutation of amino acids 172 and 174, L region), 5′-GCAGCTGGGGCCACAGGTGGCCGTAGGTGTCACG and 5′-GGCCCCAGCTGCCGCCTGGAGATGTACGAATGTGG (resulting in mutation of amino acids 199, 200, and 202, M region), and 5′-GCCGCTGCCGCCGGCACCTTTAAGCTGCCG and 5′-GGCAGCGGCCACGTGACACCTACGGCC (resulting in mutation of amino acids 211 to 213, R region). Successive mutations were introduced to generate the double (EBNA-3ALM, -3ALR, and -3AMR) and triple (EBNA-3ALMR) mutants. All mutations were confirmed by sequencing.
To express Gal4 DNA-binding domain (Gal4DBD) fusion proteins, we used the pM vector system (51). Using the Quickchange site-directed mutagenesis kit, a BamHI site was introduced into pSG5-EBNA3A at the following sites relative to the first coding nucleotide: −6 (primers 5′-CCGTCTCCTTTAAGGATCCATGGACAAGG and 5′-CCTTGTCCATGGATCCTTAAAGGAGACGG), 667 (primers 5′-GCCGCGATGTGGATCCGGGGATCGCC and 5′-GGCGATCCCCGGATCCACATCGCGGC), and 1701 (primers 5′-GCCCCGGGGATCCTTCTGGCATTAGACGC and 5′-GCGTCTAATGCCAGAAGGATCCCCGGGGC). We cloned the BamHI restriction fragments −6 to 373, 374 to 667, and 668 to 1701 into pM2 BamHI site to generate the fusion proteins Gal4DBD-3A1–126, Gal4DBD-3A125–222, and Gal4DBD-3A224–566, respectively. Using the same strategy, we generated expression plasmids for Gal4DBD-3AM125–222, Gal4DBD-3ALR125–222, and Gal4DBD-3ALMR125–222. The generation of the expression plasmids for Gal4DBD-3A455–627 and Gal4DBD-3A627–805 will be described elsewhere (D. R. Marshall, R. Dalbiès-Tran, E. Stigger-Rosser, and C. E. Sample, manuscript in preparation). Finally, we introduced a BamHI site into HVP-3A gene sequence at position 753 (primers 5′-CCATGGGTAGGATCCCGTGCCGATT and 5′-AATCGGCACGGGATCCTACCCATGG) and cloned the BamHI restriction fragment 463 to 753 into pM2 BamHI site to express Gal4DBD-HVP-3A127–223.
EBNA-3A cDNA restriction fragments (see above for details) were cloned into pGEX2T (Amersham-Pharmacia Biotech) to express glutathione S-transferase (GST)-3A125–222, GST-3A224–566, and GST-3A627–805.
Transfection and reporter assay.
A total of eight million cells were transfected by electroporation at 250 V and at 950 μF. At 48 h posttransfection, cells were harvested, extracts were prepared, and chloramphenicol acetyltransferase (CAT) activity was determined as previously described (37). The human growth hormone, used as a control for transfection efficiency, was quantitated using the HGH-TGES Radioimmunoassay Kit (Nichols Institute Diagnostics) according to the manufacturer's protocol.
Recruitment of Jκ-VP16 to the reporter promoter.
Louckes cells were transfected with 7.5 μg of pG5Ec (a CAT reporter gene under control of Gal4-binding sites 50), 1 μg pCMVhGH as a control for transfection efficiency, 5 μg of pSG5 or pSG5-JκVP16, and 5 μg of pM2 or a pM-derived plasmid.
Repression of EBNA-2-mediated transactivation.
BJAB cells were transfected with 5 μg of C10BLCAT (a CAT reporter gene under control of Jκ consensus sequences), 1 μg of pCMVhGH, 2.5 μg of pSG5-EBNA2 (or pSG5 as a control), and 2.5, 5, or 10 μg of an expression plasmid for wild-type or mutant EBNA-3A or HVP-3A (or the empty vector as a control).
Coimmunoprecipitation.
BJAB cells were cotransfected with 7.5 μg of expression plasmids for Jκ containing a hemagglutinin (HA) epitope, wild-type or mutant EBNA-3A, or the empty vector. At 48 h posttransfection, cells were harvested and lysed in 20 mM Tris (pH 8.0)–120 mM NaCl–1 mM EDTA–0.5% Nonidet P-40. Aliquots of the supernatant fraction were saved for analysis of protein expression, and the remainder was divided into two and immunoprecipitated with either an anti-HA antibody (Roche) or an anti-EBNA-3A polyclonal antibody (Exalpha Corporation, Roxbury, Mass.), followed by treatment with protein G- or protein A-Sepharose beads (Amersham-Pharmacia Biotech), respectively.
Immunoblotting.
Proteins were separated on sodium dodecyl sulfate-polyacrylamide gels and detected by immunoblotting using an anti-HA, anti-EBNA-3A, anti-EBNA-2 (Dako), or anti-c-MYC Ab-1 (Oncogene Research Products) antibody, followed by enhanced chemiluminescence (ECL System; Amersham-Pharmacia Biotech).
GST fusion chromatography.
Purification of GST fusion proteins expressed in Escherichia coli and interaction with in vitro-translated proteins have been described previously (53, 63). [35S]methionine-labeled proteins were prepared using the TNT T7 Quick Coupled Transcription-Translation System (Promega).
Cloning of HVP-3A.
DNA enriched for the HVP genome was purified from S594 cells (a gift from Fred Wang, Harvard Medical School, Boston, Mass.) using a modified Hirt method, as previously described (45). After partial digestion with XhoI and EcoRI, restriction fragments were cloned into pBKCMV, and a library was generated using the ZAP Express system (Stratagene) according to the manufacturer's protocol. This library was first screened using the HindIII restriction fragment from pBKCMV-HVP3C (64). It allowed us to determine the last 26 nucleotides of the HVP-3A coding sequence. Two oligonucleotides, one annealing in this region and a second one corresponding to nucleotides 595 to 628 of EBNA-3A cDNA, were designed as primers to generate a PCR fragment, using HVP genomic DNA as a template. This fragment was used to screen the library. A pBKCMV-HVP-3A1–2769 plasmid was isolated and used to determine the sequence of HVP-3A. To generate a pSG5-based expression vector, this plasmid was digested with BglII, and the restriction fragment containing most of the HVP-3A gene was cloned between the BamHI and the BglII sites of pSG5. Into that plasmid's BglII site was inserted the duplex oligonucleotide 5′-GATC TCGAGGATCCTCAAGATGAGGAACAGAAACTCATCTCTGAAGAGG ATCTGTAA–5′-GATCTTACAGATCCTCTTCAGAGATGAGTTTCTGTTC CTCATCTTGAGGATCCTCGA, corresponding to the HVP-3A carboxy terminus, followed by a c-MYC epitope. Finally, the amino-terminal end was reconstituted by replacing the EcoRI/FseI fragment, including the initiation codon, with the annealed oligodeoxynucleotides 5′-AATTCAAAAATGGAAGAAGACCGGCCGG and 5′-CCGGTCTTCTTCCATTTTTG to generate pSG5-HVP3A.
Nucleotide sequence accession number.
The HVP-3A gene sequence has been deposited in GenBank under accession no. AF317285.
RESULTS
Amino acids 199, 200, and 202 of EBNA-3A are essential for repression of EBNA-2–Jκ-mediated transcription.
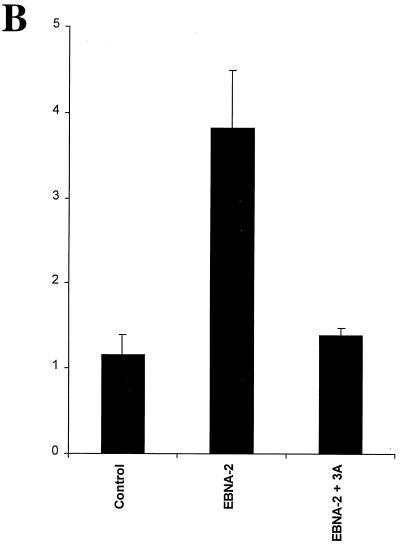
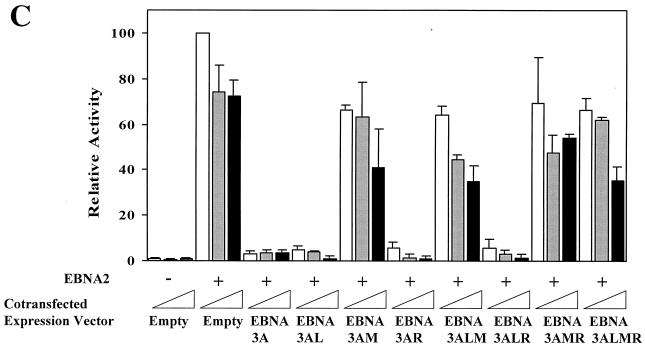
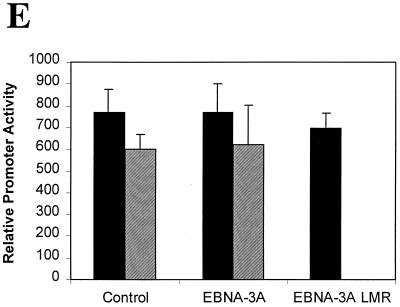
We and other groups have demonstrated that Jκ interacts with the N-proximal domain of the EBNA-3 family members (48, 63); although a few scattered amino acids can be aligned between these proteins, the domain located between amino acids 170 and 221 is the only region that has any significant homology (Fig. 1). We have generated a series of mutant EBNA-3A proteins by substituting alanine residues for the most conserved amino acids in three blocks of homologous amino acids termed L (left), M (middle), and R (right) for their location within this N-proximal conserved domain located between amino acids 170 and 221. The mutant proteins were named after the mutated regions (Fig. 1A). First, we assessed the effect of these mutations on the ability of EBNA-3A to repress EBNA-2–Jκ-mediated transcription. EBNA-3A represses transcription mediated through Jκ (6, 31), as shown here for the CD23 promoter (Fig. 1B), and has been previously shown to repress transcription mediated solely by Jκ using a synthetic promoter regulated by Jκ elements (63). Because we wished to determine the effect of these mutations specifically on Jκ-regulated transcription, we used this artificial construct in cells cotransfected with expression plasmids for EBNA-2 and various EBNA-3A proteins. As we have shown previously (reference 63 and Fig. 1B), wild-type EBNA-3A almost completely repressed transcription mediated by EBNA-2–Jκ. Mutation in either the L or the R region had no significant effect, even when both L and R mutations were present in the same molecule (EBNA-3ALR). In contrast, mutation of the M region largely prevented EBNA-3A from repressing EBNA-2–Jκ-mediated transcription, with only a slight effect exerted by a fourfold-higher level of EBNA-3AM expression vector. To ensure that EBNA-3A had no effect on EBNA-2 expression, we measured the levels of EBNA-2 protein in the presence or absence of EBNA-3A. As seen in Fig. 1D, neither the EBNA-3A wild type nor the mutant protein had any significant effect on the level of EBNA-2. Additionally, all of the EBNA-3A mutant proteins are stably expressed (data not shown). As an additional control, we examined whether EBNA-3A had any effects on constitutively active promoters not controlled by Jκ sites, such as the histone H4 and the EBV Qp promoters (Fig. 1E). EBNA-3A had no effect on expression from either of these promoters.
FIG. 1.
Amino acids 199, 200, and 202 are essential for functional interaction with Jκ. (A) Sequence homology among the EBNA-3 family members within the N-proximal region and mutations introduced into EBNA-3A are shown. Amino acids that were mutated are underlined. (B) EBNA-3A represses EBNA-2–Jκ-mediated transcription. Cells were transfected with a CD23-CAT reporter gene in the presence of EBNA-2 alone or with EBNA-3A. (C) Mutation of amino acids 199, 200, and 202 affects EBNA-3A's ability to repress EBNA-2–Jκ-mediated activation. BJAB cells were cotransfected with C10BLCAT, pCMVHGH, and pSG5-EBNA2 and with increasing amounts (2.5, 5, and 10 μg) of expression plasmids for wild-type or mutant EBNA-3A. In the controls, the empty vector pSG5 was used to maintain a constant amount of total DNA transfected. The average result and standard error of triplicate experiments are presented. (D) EBNA-3A does not affect levels of EBNA-2. Protein extracts from cells transiently transfected with EBNA-2 alone (control) or in the presence of wild-type or mutant EBNA-3A were analyzed by immunoblotting with a monoclonal antibody to EBNA-2. (E) EBNA-3A does not affect Jκ-independent transcription. Reporter genes controlled by either the histone H4 promoter (solid bars) or the EBV Qp promoter (hatched bars) were transfected into cells in the presence of wild-type or mutant EBNA-3A.
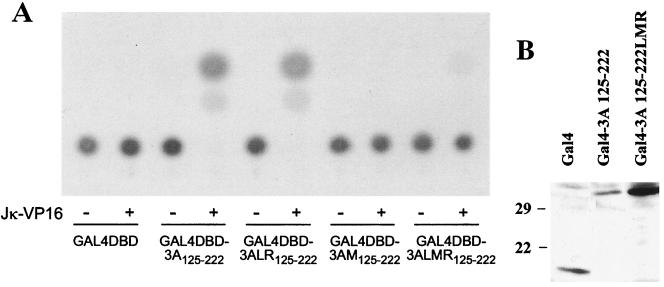
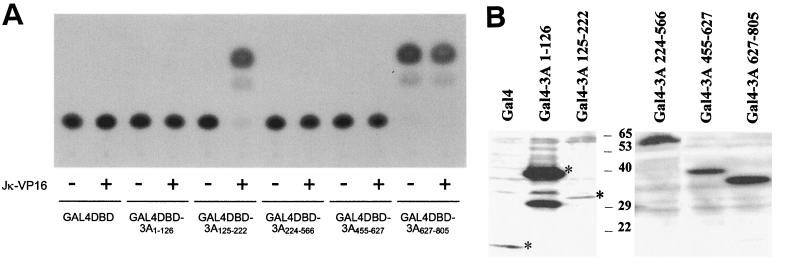
These findings indicated that these conserved amino acids were playing a significant role in the interaction of EBNA-3A with Jκ. To determine whether this domain of EBNA-3A alone is sufficient to mediate an interaction with Jκ, we performed a mammalian two-hybrid assay using amino acids 125 to 222 of wild-type or mutant EBNA-3A fused to the Gal4DBD and an expression vector for JκVP16, a fusion protein composed of Jκ and the activation domain of the herpes simplex virus transcriptional activator VP16. Interaction between these proteins was measured using a reporter gene expressed under the control of Gal4 binding sites. As seen in Fig. 2A, amino acids 125 to 222 of EBNA-3A were sufficient to interact with Jκ. Mutation in both L and R regions had no effect, whereas mutation in the M region, as well as the triple mutation, completely abolished expression of the reporter gene. As controls, we verified that the mutant protein was expressed by immunoblotting (Fig. 2B) and that the LMR mutation has no effect on the ability of the fusion protein to enter the nucleus (data not shown). These results suggested that mutation of amino acids 199, 200, and 202 in the M region prevented interaction of Jκ-VP16 with the N-proximal conserved domain of EBNA-3A.
FIG. 2.
Mutation of amino acids 199, 200, and 202 abolishes recruitment of Jκ by the N-proximal domain. (A) Louckes cells were cotransfected with pG5Ec, pCMVHGH, pSG5-JκVP16, and expression plasmids for Gal4DBD alone or fused to amino acids 125 to 222 of wild-type or mutant EBNA-3A. The data shown are representative of two experiments performed in duplicate. (B) Extracts from cells expressing Gal4DBD alone (Gal4) or fused to amino acids 125 to 222 of wild-type or LMR mutant EBNA-3A were analyzed by immunoblotting using a monoclonal antibody to Gal4.
Wild-type and mutant EBNA-3A coimmunoprecipitate with Jκ.
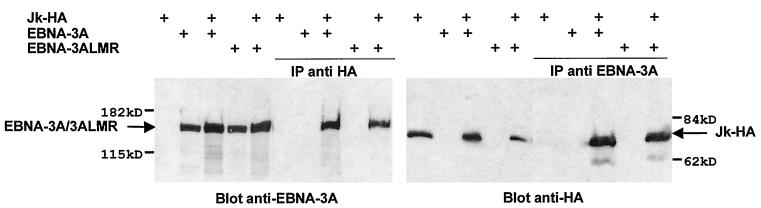
Because this mutation also significantly reduced the ability of the full-length protein to repress EBNA-2-mediated transactivation, these data suggest that these amino acids play an important role in the interaction of EBNA-3A with Jκ. To examine directly whether mutation in the M region affected the ability of EBNA-3A to interact with Jκ, we performed coimmunoprecipitation from extracts of cells transiently transfected with the expression plasmids for both Jκ tagged with an HA epitope (Jκ-HA) and wild-type or mutant EBNA-3A. As we have reported previously (63), interaction between Jκ and wild-type EBNA-3A is readily demonstrated by coimmunoprecipitation (Fig. 3). Mutation of the L and R regions, singly or in combination, had no effect on this interaction (data not shown). To our surprise however, mutation of the M region, which our other data suggested abrogated the interaction, had no effect on the interaction of the full-length protein with Jκ (Fig. 3).
FIG. 3.
Jκ and either wild-type or mutant EBNA-3A coimmunoprecipitate. BJAB cells were cotransfected with expression plasmids for Jκ-HA and/or wild-type or mutant EBNA-3A and/or the empty vector as indicated above each lane. Whole-cell extracts, present on the left of each immunoblot, were immunoprecipitated with an anti-HA or anti-EBNA-3A antibody; bound proteins were analyzed by immunoblot using anti-EBNA-3A (left panel) or anti-HA (right panel) antibodies. The migration of molecular mass markers is indicated.
Only the N-proximal domain is sufficient to interact with Jκ.
One possibility suggested by these results is that a second domain of EBNA-3A can also bind to Jκ. In support of this possibility, there have been conflicting data regarding the region of EBNA-3A that binds Jκ (3, 6, 48, 63). To resolve this issue, we sought to determine whether any other domain of EBNA-3A was sufficient to interact with Jκ by generating fusion proteins between various regions of EBNA-3A and Gal4DBD and then testing their abilities to recruit Jκ-VP16 to a Gal4-controlled promoter (Fig. 4A). The expression of all fusion proteins was verified by immunoblotting (Fig. 4B), and all were shown to localize in the nucleus by immunofluorescence with a Gal4-specific antibody (data not shown). Specific activation in the presence of Jκ-VP16 is observed only when Gal4DBD is fused to amino acids 125 to 222. In contrast, for regions 1 to 126, 224 to 566, and 455 to 627, CAT activity is limited to background level, indicating that these regions of EBNA-3A are unable to interact with Jκ. Our finding that the region between amino acids 224 and 566 did not bind Jκ is in agreement with data published by Bourillot et al. (3) but contrasts with conclusions from a GST-Jκ binding assay done by Cludts and Farrell (6).
FIG. 4.
Recruitment of Jκ by various region of EBNA-3A. (A) Louckes cells were cotransfected with pG5Ec, pCMVhGH, pSG5-JκVP16, and an expression plasmid for Gal4DBD alone or fused to the indicated regions of EBNA-3A. The data shown are representative of two experiments performed in duplicate. (B) Extracts of cells expressing Gal4DBD alone or fused to the indicated regions of EBNA-3A were analyzed by immunoblotting using a monoclonal antibody to Gal4. Asterisks indicate the positions of full-length proteins.
Because amino acids 627 to 805 constitute an activation domain (37), this assay did not allow us to conclude whether this region interacted with Jκ. To address whether this C-terminal region binds Jκ, fusion proteins between GST and EBNA-3A amino acids 125 to 222, 224 to 566, and 627 to 805 were assayed for interaction with in vitro-translated radiolabeled Jκ. Binding was weak even with GST-3A125–222 (<1% of input Jκ), and detection was rendered difficult by nonspecific binding with GST. However, no significant specific interaction between Jκ and either GST-3A224–566 or GST-3A627–805 could be detected (data not shown). Therefore, although a weak interaction with these regions is possible, we think it unlikely that they interact with Jκ.
Sequence homology between EBNA-3A and HVP-3A.
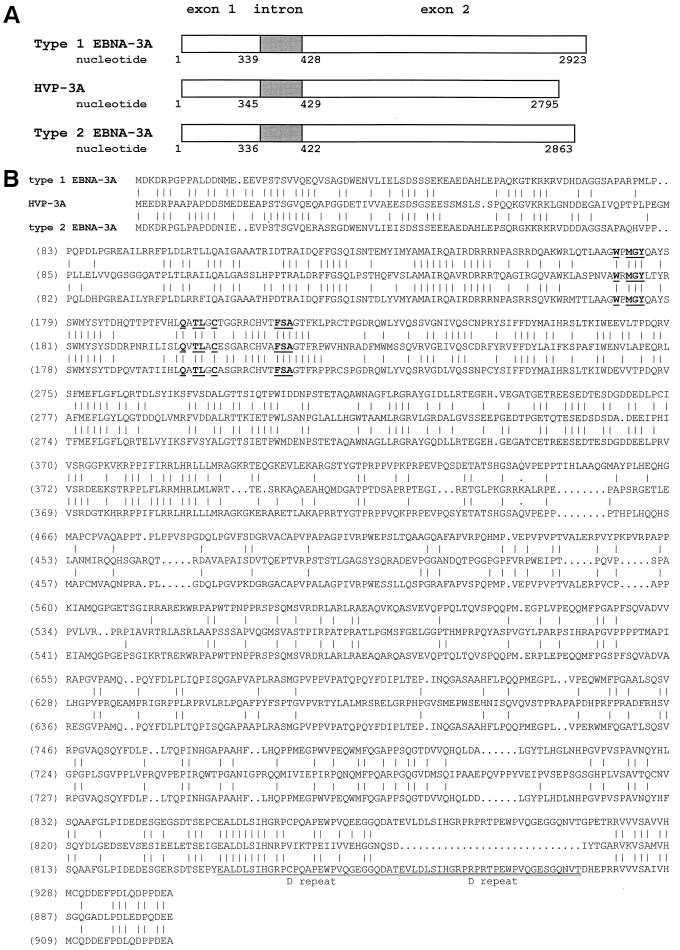
To determine the biological relevance of EBNA-3A's association with Jκ, we sought to determine whether this interaction is conserved in HVP-3A, the homolog encoded by the simian lymphocryptovirus HVP. To first determine whether the sequences that interacted with Jκ were conserved, we isolated viral genomic DNA from S594 cells and cloned and sequenced the gene encoding HVP-3A. In the 2,795-bp HVP-3A gene, we identified an intron located between nucleotides 345 and 429 which separates a short and a long exon, a gene structure that is almost identical to that of EBNA-3A (Fig. 5A). The predicted protein encoded by this gene is 903 amino acids long, slightly shorter than EBV type 1 and 2 EBNA-3A. Accordingly, its apparent molecular mass, estimated from its migration on a polyacrylamide gel under denaturing conditions (not shown), is 130 kDa compared to 160 kDa for EBNA-3A. Sequence alignment of HVP-3A and type 1 and type 2 EBNA-3A (Fig. 5B) revealed 37% identity (45% similarity) between the HVP and EBV proteins. This homology is localized mainly in the amino-terminal half, which includes the region that the above assays indicate constitutes the Jκ binding site, and at the immediate carboxy terminus. Between these two regions, the amino acid sequences are largely divergent, although both are relatively rich in glutamine and proline residues, as are the C termini of all the EBNA-3 family members.
FIG. 5.
Homology between EBV types 1 and 2 and HVP nuclear antigens 3A. (A) Gene structure. (B) Sequence alignment of the proteins. Amino acids conserved in EBNA-3 proteins are shown in boldface. The D repeat is underlined.
HVP-3A interacts with Jκ.
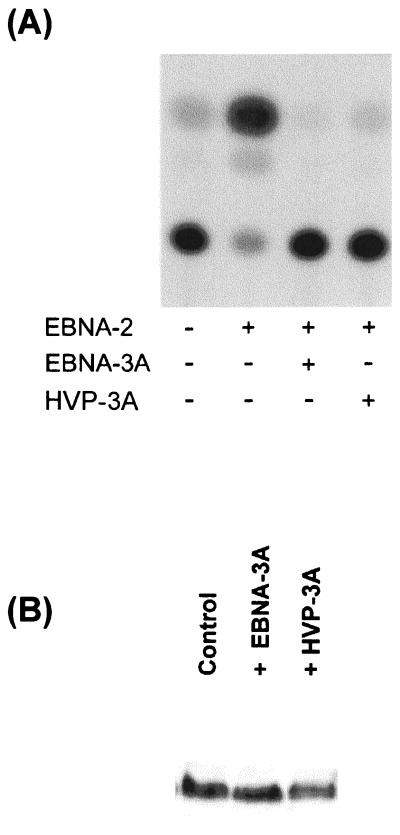
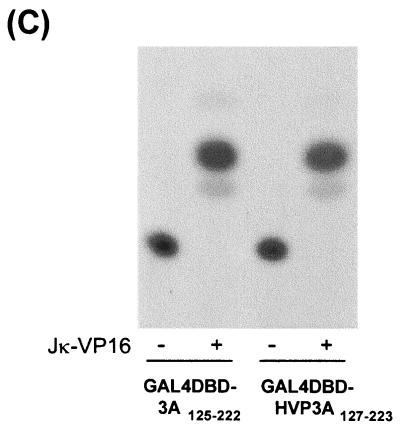
Because the domain that mediates the interaction between EBNA-3A and Jκ, including the M region, is conserved (Fig. 5B), we asked whether HVP-3A also interacts with Jκ. Because Jκ has been highly conserved during evolution, we assumed that we could use human cell lines and the human Jκ gene in reporter assays identical to those described above. Like EBNA-3A, HVP-3A was able to repress EBNA-2-mediated transcription activation (Fig. 6A). Immunoblotting confirmed that HVP-3A, like EBNA-3A, had no effect on expression of EBNA-2 (Fig. 5B). To confirm that HVP-3A interacts with Jκ through the corresponding domain, we fused the Gal4DBD to amino acids 127 to 223 of HVP-3A. A strong and specific activation of the CAT gene was detected only in presence of Jκ-VP16 (Fig. 6C), demonstrating its recruitment to the promoter. These results indicate that throughout evolution nuclear antigen 3A has conserved its ability to repress EBNA-2-mediated transactivation and to interact with the cellular transcription factor Jκ through a domain proximal to the amino terminus.
FIG. 6.
Interaction with Jκ is conserved in HVP-3A. (A) HVP-3A represses EBNA-2–Jκ-mediated activation. BJAB cells were cotransfected with C10BLCAT, pCMVHGH, pSG5-EBNA2, and expression plasmids for EBV or HVP nuclear antigen 3A, or pSG5 in control experiments. The data shown are representative of three experiments performed in duplicate. (B) Extracts from cells transfected with EBNA-2 alone (control) or in the presence of EBNA-3A or p3A proteins were analyzed by immunoblotting using a monoclonal antibody to EBNA-2. (C) HVP-3A N-proximal region recruits Jκ. Louckes cells were cotransfected with pG5Ec, pCMVhGH, and expression plasmids for Gal4DBD alone or fused to the N-proximal region of EBNA-3A or HVP-3A. The data shown are representative of two experiments performed in duplicate.
DISCUSSION
Of more than 80 genes potentially encoded by the EBV genome, only 12 are expressed during latent infection in vitro. This restricted pattern of expression is possible because the virus harnesses the activity of numerous cellular proteins. Not only does EBV use the cellular transcriptional machinery to express the LMP, BARF0, or EBNA genes, but viral proteins in turn cooperate with cellular factors to exert their activity. A well-studied example is EBNA-2, a transcriptional activator that is targeted to its responsive promoters through interaction with Jκ, a ubiquitous sequence-specific DNA-binding transcription factor. The Jκ consensus sequence is present within the promoters of many cellular and viral genes, including EBV latent promoters. Proteins of the EBNA-3 family can also interact with Jκ and repress EBNA-2–Jκ-mediated transcriptional activation in reporter gene assays (25, 30, 32, 37, 47, 48, 58, 61, 63). It has been suggested that interaction between Jκ and the EBNA-2 and -3 proteins may be involved in the upregulation of interleukin-1β expression in EBV-infected cells (29). Together, these data position Jκ at the heart of the dialog between the virus and its host during latent infection.
In this study, we have characterized the association between EBNA-3A and Jκ, and demonstrated its conservation in the simian HVP homolog. EBNA-3A interacts with Jκ through a N-proximal region located between amino acids 125 and 222. This is the only region of significant sequence homology between the EBNA-3 family members, and it is contained within the domain from amino acids 1 to 223 that was originally reported by our group to bind Jκ as demonstrated by an in vitro experimental approach (63). Since subsequent studies provided conflicting data as to the domain of EBNA-3A that was involved (3, 6, 48), we reexamined this question using a sensitive assay that would detect interaction within the cell. We demonstrate (i) that the N-proximal domain, located between amino acids 125 and 222, is sufficient to recruit Jκ in B cells and (ii) by site-directed mutagenesis, that residues 199, 200, and 202 within this domain are of particular importance in this interaction and in the ability of EBNA-3A to regulate transcription mediated through Jκ. In contrast, mutation of other short sequences conserved among the EBNA-3 proteins (i.e., amino acids 172, 174, and 211 to 213 of EBNA-3A) had no detectable consequence in the same experiments. Surprisingly, the loss of transcriptional regulation by EBNA-3A does not correlate with its inability to interact with Jκ, since the full-length EBNA-3AM remained able to coimmunoprecipitate with Jκ. Our first hypothesis was that a second region of EBNA-3A was also able to bind Jκ. In support of this hypothesis, others have reported a direct interaction with the amino-terminal 138 amino acids (48) or the central region (residues 224 to 566) (6), but these conclusions were based on a weak interaction between in vitro-translated and GST fusion proteins. Our results show that no region outside the N-proximal conserved domain is sufficient to mediate recruitment of Jκ in B cells, although we cannot exclude that such a region might have a role in stabilizing the association in the context of the whole molecule.
One possibility that may explain our results is that EBNA-3A and Jκ are components of a higher-order protein complex. Since previous data favor a direct contact between the two proteins (63), we propose that within such a complex, the N-proximal region of EBNA-3A interacts with Jκ but that one or several cofactors stabilize the association. This model would also explain why the interaction observed in vitro appears weak, while the interaction is readily seen in vivo (63), although an alternative explanation is that posttranslational modifications are required for an efficient interaction. Consistent with this hypothesis, EBNA-3 proteins, as well as Jκ, have been recently reported to associate with multiprotein complexes involved in transcription, such as the RNA polymerase II holoenzyme (40), a histone deacetylase complex (21, 27, 44) as well as cellular transcription factors (54, 64). It is therefore conceivable that mutation of residues 199, 200, and 202 does not disrupt the whole complex but rather generates sufficient instability that transcription mediated through Jκ is affected.
There are two major subtypes of EBV that exhibit differences mostly in the EBNA-2 and -3 proteins. The identities between type 1 and 2 proteins were 54, 84, 80, and 72% for EBNA-2, -3A, -3B, and -3C, respectively (7, 52). In the context of this study, it is important to note that EBNA-3A and -3C encoded by both EBV subtypes have been shown to interact with Jκ in B cells (30, 58). Although the HVP-3A gene exhibits a structural homology with EBNA-3A with a short and a long exon, the amino acid sequence is relatively divergent. Sequence alignment of HVP-3A and type 1 and 2 EBNA-3A reveals 37% identity (45% homology) in the predicted amino acid sequences (Fig. 4B), values within the range observed for the other latent proteins. EBNA-2 types 1 and 2 are 35 and 37% identical, respectively, to their homologs encoded by HVP extracted from B65 cells, and the simian virus protein did not appear to fall in either category (36). HVP-3A and EBNA-3A types 1 and 2 have predicted lengths of 903, 944, and 925 amino acids, respectively. Most of the difference is accounted for by a 29-amino-acid element, the so-called D repeat, near the carboxy terminus; HVP-3A contains only one D repeat, whereas EBNA-3A contains two. Conservation of this sequence suggests that it probably has functional importance, which remains to be characterized, while the presence of a single copy in HVP-3A implies that its repetition in EBNA-3A is simply redundant. Shorter deletions or insertions of one to six amino acids can be found in HVP-3A in regions that are well conserved between the EBV subtypes but, interestingly, none of the insertions characteristic of type 1 EBNA-3A are present in HVP-3A. This suggests that the HVP isolated from S594 cells is closer to type 2 EBV. To our knowledge, there is to date no evidence for different subtypes of HVP, but the existence of two subtypes of the related rhesus virus has been recently reported (5). Given that several features of the phylogenetic tree of herpesviruses support the hypothesis of cospeciation with the host (for a review, see reference 38), it is tempting to assume that divergence into two subtypes is anterior to differentiation between Old World primates and hominidae. Nevertheless, the possibility of cross-species infection and recombination of one subtype with another virus, most likely a primate virus, exists. Sequencing of EBNA-2, -3A, -3B, and -3C homologs from different strains of HVP and rhesus lymphocryptoviruses may further support one of these hypotheses. The identity between EBNA-3A and HVP-3A is mainly localized to the amino-terminal half, a finding reminiscent of the homology among the EBNA-3 proteins. Amino acids that we characterized as essential in the M region are conserved in HVP-3A and, like EBNA-3A, the N-proximal domain between amino acids 127 and 223 interacts with human Jκ. These findings are consistent with our observations that these residues play an essential role in the interaction of EBNA-3A with Jκ. Conservation throughout evolution of the interaction and resulting transcriptional regulatory activity suggests that they play a significant biological role.
Interaction with Jκ is a common feature of the EBNA-3 family members and involves a conserved domain of these proteins. Yet our group has demonstrated that the mutant EBNA-3CLMR protein and Jκ do not coimmunoprecipitate when overexpressed in B cells (Marshall et al., in preparation), in contrast to the observation published here for EBNA-3ALMR. Moreover, mutations in both the L and R regions altered EBNA-3C's ability to repress Jκ-mediated activation of transcription (Marshall et al., in preparation), whereas EBNA-3ALR retained this ability to repress transcription. This might reflect a difference in the interaction of EBNA-3A and EBNA-3C with Jκ itself or, more likely, with different cofactors within multiprotein complexes and raises the possibility that interaction of EBNA-3A and EBNA-3C with Jκ influence distinct molecular pathways. Based on our results, recombinant EBV encoding mutant EBNA-3A can now be generated and used to investigate whether this transcriptional regulatory mechanism is essential for the EBV-mediated immortalization of B lymphocytes in vitro.
ACKNOWLEDGMENTS
We thank Fred Wang (Harvard Medical School, Boston) for the gift of the S594 cell line; Patricia Vaughan and Gary Stein (University of Massachusetts, Worcester) and Jeff Sample for the histone H4 and Qp reporter constructs, respectively; Bruno Chatton (IGBMC, Strasbourg, France) for the Gal4 antibody; and Bo Zhao and Mikhail Matrosovich for helpful discussions.
This research was supported by Public Health Service grant CA73561 and CA56645, Cancer Center Support (CORE) grant CA21765, and the American Lebanese Syrian Associated Charities (ALSAC).
REFERENCES
- 1.Arvanis-Tsakonas S, Rand M D, Lake R J. Notch signalling: cell fate control and signal integration in development. Science. 1999;284:770–776. doi: 10.1126/science.284.5415.770. [DOI] [PubMed] [Google Scholar]
- 2.Bailey A M, Posakony J W. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes Dev. 1995;9:2609–2622. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- 3.Bourillot P, Waltzer L, Sergeant A, Manet E. Transcriptional repression by the Epstein Barr virus EBNA-3A protein tethered to DNA does not require RBP-Jκ. J Gen Virol. 1998;79:363–370. doi: 10.1099/0022-1317-79-2-363. [DOI] [PubMed] [Google Scholar]
- 4.Brou C, Logeat F, Lecourtois M, Vandekerckhove J, Kourilsky P, Schweisguth F, Israel A. Inhibition of the DNA-binding activity of Drosophila Suppressor of Hairless and of its human homolog, KBF2/RBP-Jκ, by direct protein-protein interaction with Drosophila Hairless. Genes Dev. 1994;8:2491–2503. doi: 10.1101/gad.8.20.2491. [DOI] [PubMed] [Google Scholar]
- 5.Cho Y G, Gordadze A V, Ling P D, Wang F. Evolution of two types of rhesus lymphocryptovirus similar to type 1 and type 2 Epstein-Barr virus. J Virol. 1999;73:9206–9212. doi: 10.1128/jvi.73.11.9206-9212.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cludts I, Farrell P J. Multiple functions within the Epstein-Barr virus EBNA-3A protein. J Virol. 1998;72:1862–1869. doi: 10.1128/jvi.72.3.1862-1869.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dambaugh T, Hennessy K, Chamnankit L, Kieff E. U2 region of the Epstein-Barr virus DNA may encode Epstein-Barr virus nuclear antigen 2. Proc Natl Acad Sci USA. 1984;81:7632–7636. doi: 10.1073/pnas.81.23.7632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dou S, Zeng X, Cortes P, Erdjument-Bromage H, Tempst P, Honjo T, Vales L D. The recombination signal sequence-binding protein RBP-2N functions as a transcriptional repressor. Mol Cell Biol. 1994;14:3310–3319. doi: 10.1128/mcb.14.5.3310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falk L, Deinhardt F, Nonoyama M, Wolfe L G, Bergholz C. Properties of a baboon lymphotropic herpesvirus related to Epstein-Barr virus. Int J Cancer. 1976;18:798–807. doi: 10.1002/ijc.2910180611. [DOI] [PubMed] [Google Scholar]
- 10.Franken M, Annis B, Ali A N, Wang F. 5′ Coding and regulatory sequence divergence with conserved function of the Epstein-Barr virus LMP2A homolog in herpesvirus papio. J Virol. 1995;69:8011–8019. doi: 10.1128/jvi.69.12.8011-8019.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Franken M, Devergne O, Rosenzweig M, Annis B, Kieff E, Wang F. Comparative analysis identifies conserved tumor necrosis factor receptor-associated factor 3 binding sites in the human and simian Epstein-Barr virus oncogene LMP1. J Virol. 1996;70:7819–7826. doi: 10.1128/jvi.70.11.7819-7826.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fries K L, Sculley T B, Webster-Cyriaque J, Rajadurai P, Sadler R H, Raab-Traub N. Identification of a novel protein encoded by the BamHI A region of the Epstein-Barr virus. J Virol. 1997;71:2765–2771. doi: 10.1128/jvi.71.4.2765-2771.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuentes-Panama E M, Swaminathan S, Ling P. Transcriptional activation signals found in the Epstein-Barr virus (EBV) latency C promoter are conserved in the latency C promoter sequences from baboon and rhesus monkey EBV-like lymphocryptoviruses (cercopithicine herpesviruses 12 and 15) J Virol. 1999;73:826–833. doi: 10.1128/jvi.73.1.826-833.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerber P, Kalter S S, Schidlovsky G, Peterson W D, Daniel M D. Biologic and antigenic characteristics of Epstein-Barr virus-related herpesviruses of chimpanzees and baboons. Int J Cancer. 1977;20:448–459. doi: 10.1002/ijc.2910200318. [DOI] [PubMed] [Google Scholar]
- 15.Grossman S R, Johannsen E, Tong X, Yalamanchili R, Kieff E. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the Jκ recombination signal binding protein. Proc Natl Acad Sci USA. 1994;91:7568–7572. doi: 10.1073/pnas.91.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heller M, Kieff E. Colinearity between the DNAs of Epstein-Barr virus and herpesvirus papio. J Virol. 1981;37:821–826. doi: 10.1128/jvi.37.2.821-826.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Henkel T, Ling P D, Hayward S D, Peterson M G. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science. 1994;265:92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- 18.Howe J G, Shu M D. Isolation and characterization of the genes for two small RNAs of herpesvirus papio and their comparison with Epstein-Barr virus-encoded EBER RNAs. J Virol. 1988;62:2790–2798. doi: 10.1128/jvi.62.8.2790-2798.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hsieh J J, Hayward S D. Masking of the CBF1/RBPJ kappa transcriptional repression domain by Epstein-Barr virus EBNA2. Science. 1995;268:560–563. doi: 10.1126/science.7725102. [DOI] [PubMed] [Google Scholar]
- 20.Hsieh J J, Henkel T, Salmon P, Robey E, Peterson M G, Hayward S D. Truncated mammalian Notch1 activates CBF1/RBPJκ-repressed genes by a mechanism resembling that of Epstein-Barr virus EBNA2. Mol Cell Biol. 1996;16:952–959. doi: 10.1128/mcb.16.3.952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsieh J J, Zhou S, Chen L, Young D B, Hayward S D. CIR, a corepressor linking the DNA binding factor CBF1 to the histone deacetylase complex. Proc Natl Acad Sci USA. 1999;96:23–28. doi: 10.1073/pnas.96.1.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jarriault S, Brou C, Logeat F, Schroeter E H, Kopan R, Israël A. Signaling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 23.Jarriault S, Le Bail O, Hirsinger E, Pourquie O, Logeat F, Strong C F, Brou C, Seidah N G, Israël A. delta-1 activation of Notch-1 signalling results in HES-1 transactivation. Mol Cell Biol. 1998;18:7423–7431. doi: 10.1128/mcb.18.12.7423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johannsen E, Koh E, Mosialos G, Tong X, Kieff E, Grossman S R. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by Jκ and PU.1. J Virol. 1995;69:253–262. doi: 10.1128/jvi.69.1.253-262.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johannsen E, Miller C L, Grossman S R, Kieff E. EBNA 2 and EBNA 3C extensively and mutually exclusively associate with RBPJκ in Epstein-Barr virus-transformed B lymphocytes. J Virol. 1996;70:4179–4183. doi: 10.1128/jvi.70.6.4179-4183.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kannabiran C, Zeng X, Vales L D. The mammalian transcriptional repressor RBP (CBF1) regulates interleukin-6 gene expression. Mol Cell Biol. 1997;17:1–9. doi: 10.1128/mcb.17.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kao H Y, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner C R, Evans R M, Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Raven Press; 1996. pp. 2343–2396. [Google Scholar]
- 29.Krauer K G, Belzer D K, Liaskou D, Buck M, Cross S, Honjo T, Sculley T. Regulation of interleukin-1β transcription by Epstein-Barr virus involves a number of latent proteins via their interaction with RBP. Virology. 1998;252:418–430. doi: 10.1006/viro.1998.9441. [DOI] [PubMed] [Google Scholar]
- 30.Krauer K G, Kienzle N, Young S B, Sculley T B. Epstein-Barr nuclear antigen-3 and -4 interact with RBP-2N, a major isoform of RBP-Jκ in B lymphocytes. Virology. 1996;226:346–353. doi: 10.1006/viro.1996.0662. [DOI] [PubMed] [Google Scholar]
- 31.Laux G, Dugrillon F, Eckert C, Adam B, Zimber-Strobl U, Bornkamm G W. Identification and characterization of an Epstein-Barr virus nuclear antigen 2-responsive cis element in the bidirectional promoter region of latent membrane protein and terminal protein 2 genes. J Virol. 1994;68:6947–6958. doi: 10.1128/jvi.68.11.6947-6958.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Le Roux R A, Kerdiles B, Walls D, Dedieu J F, Perricaudet M. The Epstein-Barr virus determined nuclear antigens EBNA 3A, 3B, and 3C repress EBNA 2-mediated transactivation of the viral terminal protein 1 gene promoter. Virology. 1994;205:596–602. doi: 10.1006/viro.1994.1687. [DOI] [PubMed] [Google Scholar]
- 33.Lecourtois M, Schweisguth F. The neurogenic Suppressor of Hairless DNA-binding protein mediates the transcriptional activation of the Enhancer of split complex genes triggered by notch signaling. Genes Dev. 1995;9:2598–2608. doi: 10.1101/gad.9.21.2598. [DOI] [PubMed] [Google Scholar]
- 34.Ling P D, Hayward S D. Contribution of conserved amino acids in mediating the interaction between EBNA2 and CBF1/RBPJκ. J Virol. 1995;69:1944–1950. doi: 10.1128/jvi.69.3.1944-1950.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ling P D, Rawlins D R, Hayward S D. The Epstein-Barr virus immortalizing protein EBNA 2 is targeted to DNA by a cellular enhancer-binding protein. Proc Natl Acad Sci USA. 1993;90:9237–9241. doi: 10.1073/pnas.90.20.9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ling P D, Ryon J J, Hayward S D. EBNA-2 of herpesvirus papio diverges significantly from the type A and type B EBNA-2 proteins of Epstein-Barr virus but retains an efficient transactivation domain with a conserved hydrophobic motif. J Virol. 1993;67:2990–3003. doi: 10.1128/jvi.67.6.2990-3003.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Marshall D R, Sample C E. Epstein-Barr virus nuclear antigen 3C is a transcriptional regulator. J Virol. 1995;69:3624–3630. doi: 10.1128/jvi.69.6.3624-3630.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McGeoch D J, Davison A J. The molecular evolutionary history of the herpesviruses. In: Domingo E, Webster R G, Holland J, editors. Origin and evolution of viruses. London, United Kingdom: Academic Press; 1999. pp. 441–465. [Google Scholar]
- 39.Moghaddam A, Rosenzweig M, Lee-Parritz D, Annis B, Johnson R P, Wang F. An animal model for acute and persistent Epstein-Barr virus infection. Science. 1997;276:2030–2033. doi: 10.1126/science.276.5321.2030. [DOI] [PubMed] [Google Scholar]
- 40.Olave I, Reinberg D, Vales L D. The mammalian transcriptional repressor RBP (CBF1) targets TFIID and TFIIA to prevent activated transcription. Genes Dev. 1998;12:1621–1637. doi: 10.1101/gad.12.11.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oswald F, Liptay S, Adler G, Schmid R. NF-κB2 is a putative target gene of activated Notch-1 via RBP-Jκ. Mol Cell Biol. 1998;18:2077–2088. doi: 10.1128/mcb.18.4.2077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peng R S, Gordadze A V, Fuentes-Panama E M, Wang F, Zong J, Hayward G S, Tan J, Ling P D. Sequence and functional analysis of EBNA-LP and EBNA2 proteins from nonhuman primate lymphocryptoviruses. J Virol. 1999;74:379–389. doi: 10.1128/jvi.74.1.379-389.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Plaisance S, vanden Berghe W, Boone E, Friers W, Haegeman G. Recombination signal sequence binding protein Jκ is constitutively bound to the NF-κB site of the interleukin-6 promoter and acts as a negative regulatory factor. Mol Cell Biol. 1997;17:3733–3743. doi: 10.1128/mcb.17.7.3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Radkov S A, Touitou R, Brehm A, Rowe M, West M, Kouzarides T, Allday M J. Epstein-Barr virus nuclear antigen 3C interacts with histone deacetylase to repress transcription. J Virol. 1999;73:5688–5697. doi: 10.1128/jvi.73.7.5688-5697.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Redemann B E, Mendelson E, Carter B J. Adeno-associated virus Rep protein synthesis during productive infection. J Virol. 1989;63:873–882. doi: 10.1128/jvi.63.2.873-882.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rickinson A, Kieff E. Epstein-Barr virus. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Philadelphia, Pa: Raven Publishers; 1996. pp. 2397–2446. [Google Scholar]
- 47.Robertson E S, Grossman S, Johannsen E, Miller C, Lin J, Tomkinson B, Kieff E. Epstein-Barr virus nuclear protein 3C modulates transcription through interaction with the sequence-specific DNA-binding protein J kappa. J Virol. 1995;69:3108–3116. doi: 10.1128/jvi.69.5.3108-3116.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Robertson E, Lin J, Kieff E. The amino-terminal domains of Epstein-Barr virus nuclear proteins 3A, 3B, and 3C interact with RBPJκ. J Virol. 1996;70:3068–3074. doi: 10.1128/jvi.70.5.3068-3074.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ryon J J, Fixman E D, Houchens C, Zong J, Lieberman P M, Chang Y N, Hayward G S, Hayward S D. The lytic origin of herpesvirus papio is highly homologous to Epstein-Barr virus ori-Lyt: evolutionary conservation of transcriptional activation and replication signals. J Virol. 1993;67:4006–4016. doi: 10.1128/jvi.67.7.4006-4016.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sadowski I, Bell B, Broad P, Hollis M. GAL4 fusion vectors for expression in yeast or mammalian cells. Gene. 1992;118:137–141. doi: 10.1016/0378-1119(92)90261-m. [DOI] [PubMed] [Google Scholar]
- 51.Sample J T, Sample C E. Epstein-Barr virus: molecular biology. In: Granoff A, Webster R G, editors. Encyclopedia of virology. London, England: Academic Press; 1999. pp. 494–501. [Google Scholar]
- 52.Sample J T, Young L, Martin B, Chatman T, Kieff E, Rickinson A. Epstein-Barr virus types 1 and 2 differ in their EBNA 3A, EBNA 3B, and EBNA 3C genes. J Virol. 1990;64:4084–4092. doi: 10.1128/jvi.64.9.4084-4092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith D B, Johnson K S. Single-step purification of polypeptides expressed in Escherichia coli as fusions with glutathione S-transferase. Gene. 1988;67:31–40. doi: 10.1016/0378-1119(88)90005-4. [DOI] [PubMed] [Google Scholar]
- 54.Taniguchi Y, Furukawa T, Tun T, Han H, Honjo T. LIM protein KyoT2 negatively regulates transcription by association with the RBP-J DNA-binding protein. Mol Cell Biol. 1998;18:644–654. doi: 10.1128/mcb.18.1.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tomkinson B, Robertson E, Kieff E. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J Virol. 1993;67:2014–2025. doi: 10.1128/jvi.67.4.2014-2025.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tsang S, Wang F, Izumi K M, Kieff E. Delineation of the cis-acting element mediating EBNA-2 transactivation of latent infection membrane protein association. J Virol. 1991;65:6765–6771. doi: 10.1128/jvi.65.12.6765-6771.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yates J L, Camiolo S M, Ali S, Ying A. Comparison of the EBNA1 proteins of Epstein-Barr virus and herpesvirus papio in sequence and function. Virology. 1996;222:1–13. doi: 10.1006/viro.1996.0392. [DOI] [PubMed] [Google Scholar]
- 58.Young D B, Krauer K, Kienzle N, Sculley T. Both A type and B type Epstein-Barr virus nuclear antigen 6 interact with RBP-2N. J Gen Virol. 1997;78:1671–1674. doi: 10.1099/0022-1317-78-7-1671. [DOI] [PubMed] [Google Scholar]
- 59.Waltzer L, Bourillot P Y, Sergeant A, Manet E. RBP-Jκ repression activity is mediated by a co-repressor and antagonized by the Epstein-Barr virus transcription factor EBNA 2. Nucleic Acids Res. 1995;23:4939–4945. doi: 10.1093/nar/23.24.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Waltzer L, Logeat F, Brou C, Israel A, Sergeant A, Manet E. The human Jκ recombination signal sequence binding protein (RBP-Jκ) targets the Epstein-Barr virus EBNA 2 protein to its DNA responsive elements. EMBO J. 1994;13:5633–5638. doi: 10.1002/j.1460-2075.1994.tb06901.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Waltzer L, Perricaudet M, Sergeant A, Manet E. Epstein-Barr virus EBNA 3A and EBNA 3C proteins both repress RBP-Jκ-EBNA2-activated transcription by inhibiting the binding of RBP-Jκ to DNA. J Virol. 1996;70:5909–5915. doi: 10.1128/jvi.70.9.5909-5915.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang F, Tsang S, Kurilla M G, Cohen J I, Kieff E. Epstein-Barr virus nuclear antigen 2 transactivates latent membrane protein LMP-1. J Virol. 1990;64:3407–3416. doi: 10.1128/jvi.64.7.3407-3416.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhao B, Marshall D R, Sample C E. A conserved domain of the Epstein-Barr virus nuclear antigens 3A and 3C binds to a discrete domain of Jκ. J Virol. 1996;70:4228–4236. doi: 10.1128/jvi.70.7.4228-4236.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhao B, Sample C E. Epstein-Barr virus nuclear antigen 3C activates the latent membrane protein 1 promoter in the presence of Epstein-Barr virus nuclear antigen 2 through sequences encompassing an Spi-1/Spi-B binding site. J Virol. 2000;74:5151–5160. doi: 10.1128/jvi.74.11.5151-5160.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]