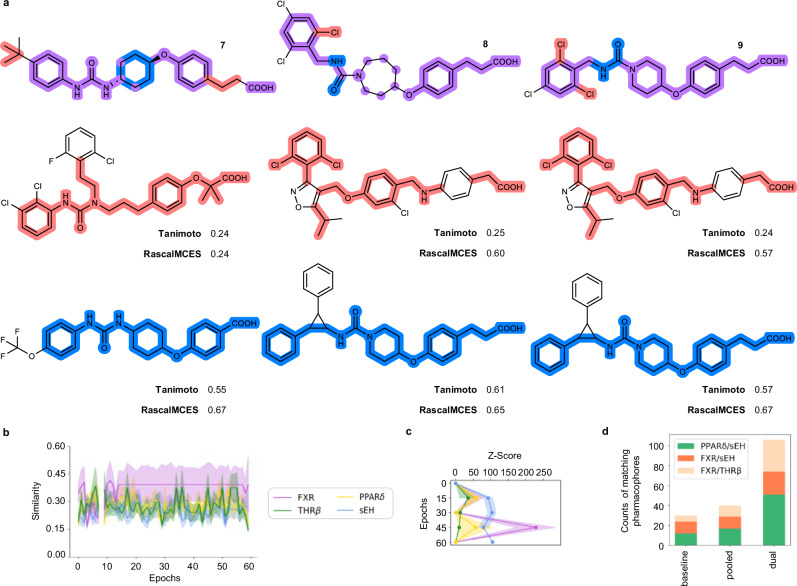
Fig. 6. Validation and comparison of the dual ligand design approach.
a Comparison of designed dual PPARδ/sEH modulators with the most similar fine-tuning templates by Tanimoto similarity computed on Morgan fingerprints and Rapid Similarity Calculation of Maximum Common Edge Subgraph (RascalMCES)45. The highlighted substructures represent the extracted RascalMCES between the most similar fine-tuning templates and the designed dual modulators (red—PPARδ, blue—sEH, violet—both). b Effects of chemical language model (CLM) fine-tuning with the pooled templates for FXR, THRβ, PPARδ and sEH. The max. Tanimoto similarity ± SD (standard deviation of max. Tanimoto similarity) computed on Morgan fingerprints of the beam search designs (width 50) per epoch to the fine-tuning molecules for the respective target is shown. For each epoch beam search designs (width 50) were generated and only valid SMILES were analyzed. c Target prediction (Z-scores) of beam search designs (width 50) over the fine-tuning procedure using the Similarity Ensemble Approach (SEA)39 for the targets of interest. Z-Scores are the mean ± SD. For each epoch beam search designs (width 50) were generated and only valid SMILES were analyzed. d Bar chart of fractions of designs obtained from the native baseline CLM, after fine-tuning with the pooled templates for FXR, THRβ, PPARδ and sEH, or the pooled ligands for the target pairs of interest (dual) matching pharmacophore models (Supplementary Fig. 4) of the respective target pairs. Source data are provided as a Source Data file.