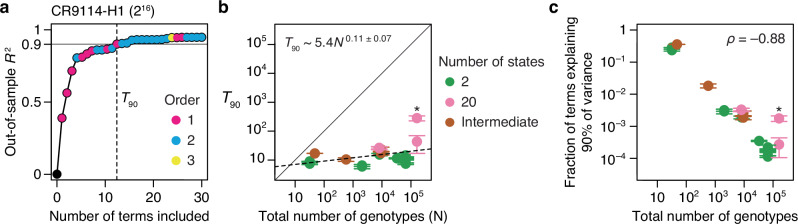
Fig. 6. Sparsity of protein sequence-function relationships.
a Measuring the sparsity of genetic architecture, illustrated using the CR9114-H1 dataset. RFA terms up to third order were estimated and ranked by the fraction of variance they explain (calculated using the simple method for computing the variance contribution of each term described in Methods). Models of increasing complexity were constructed by sequentially including each term, and each model was evaluated by cross-validation. Each dot represents a model, colored by the order of the last term added. Vertical line marks T90, the minimal number of terms required for an out-of-sample R2 of 0.9. b T90 as a function of the total number of genotypes. Dotted line, best-fit power function. Asterisk, GB1 dataset. Each T90 was estimated in two ways: as the number of terms required to reach R2 of 0.9 (upper error bar)—an overestimate because measurement noise prevents any model from attaining an out-of-sample R2 of 1—and as the number of terms required for an R2 equal to 90% of that of the full third-order model (lower error bar); circles show the average of the two estimates. c Fraction of all terms required to explain 90% of phenotypic variance shown against the total number of genotypes. Asterisk, GB1 dataset. Error bars show the possible maximum and minimum computed as in (b).