Abstract
Multiple myeloma (MM) is a hematological malignancy whose curability is greatly challenged by recurrent patient relapses and therapy resistance. We have previously proposed the high expression of ADAM8, ADAM9 and ADAM15 (A Disintegrin And Metalloproteinase 8/9/15) as adverse prognostic markers in MM. This study focused on the so far scarcely researched role of ADAM8/9/15 in MM using two patient cohorts and seven human MM cell lines (HMCL). High ADAM8/9/15 expression was associated with high-risk cytogenetic abnormalities and extramedullary disease. Furthermore, ADAM8/15 expression increased with MM progression and in relapsed/refractory MM compared to untreated patient samples. RNA sequencing and gene set enrichment analysis comparing ADAM8/9/15high/low patient samples revealed an upregulation of proliferation markers and proliferation-associated gene sets in ADAM8/9/15high patient samples. High ADAM8/9/15 expression correlated with high Ki67 and high ADAM8/15 expression with high MYC protein expression in immunohistochemical stainings of patient tissue. Conversely, siRNA-mediated knockdown of ADAM8/9/15 in HMCL downregulated proliferation-related gene sets. Western blotting revealed that ADAM8 knockdown regulated IGF1R/AKT signaling and ADAM9 knockdown decreased mTOR activation. Lastly, high ADAM8/9/15 expression levels were verified as prognostic markers independent of Ki67/MYC expression and/or high-risk abnormalities. Overall, these findings suggest that ADAM8/9/15 play a role in MM progression and proliferation signaling.
Subject terms: Oncogenes, Cell signalling, Risk factors, Myeloma
Introduction
Multiple myeloma (MM) represents approximately 10% of all hematological malignancies and patients present with a variety of clinical manifestations and diverse cytogenetic backgrounds [1]. While MM patient survival has improved due to novel treatments over the last ten years [2], the disease is still generally considered incurable with a median survival of approximately six years [3]. Therapy resistance remains as a major obstacle, causing almost all patients to eventually relapse [1]. Therefore, there is a persisting need to identify novel therapeutic targets and biomarkers associated with disease progression.
The interaction of MM cells with the bone marrow microenvironment, comprised of cellular components (hematopoietic, endothelial and bone marrow stromal cells, osteoblasts, osteocytes, fibroblasts, osteoclasts, etc.) providing growth factors, cytokines and chemokines as well as a non-cellular compartment (extracellular matrix (ECM)), is important for MM cell proliferation and disease progression [4, 5]. We have previously found an association between the high gene expression (GE) of ECM genes such as A Disintegrin and Metalloproteinase (ADAM) 8, ADAM9 and ADAM15 and a significantly shorter progression-free (PFS) and overall survival (OS) in MM patients [6].
The ADAM family comprises transmembrane and secreted proteins which are involved in various processes important for cancer [7, 8]. ADAM8, 9 and 15 are proteolytically active transmembrane proteins known to shed ectodomains of, among others, growth factors, cytokines and receptors [7, 8]. ADAMs also interact with a variety of other proteins such as integrins and affect integrin signaling pathways such as AKT and mitogen-activated protein kinase (MAPK) signaling [7, 9]. Intracellular signaling is also mediated by their cytoplasmic domains, which contain binding sites for SH3-domain-containing proteins (e.g. phosphoinositide-3-kinase (PI3K)) and amino acid residues which can be phosphorylated by kinases [7, 10].
Accordingly, the upregulation of ADAM8, ADAM9 and ADAM15 has been described, among others, in the context of prognosis, proliferation and progression of e.g. hepatocellular, renal, lung, breast, bladder and colon cancer [9–22].
In contrast, research concerning the role of ADAM8, 9 and 15 in MM is scarce [23–25] and studies thoroughly examining the influence of ADAM8/9/15 on clinical parameters and signaling pathways in MM are lacking. This study therefore aimed to gain insight into the clinical and functional role of ADAM8, ADAM9 and ADAM15 in primary MM and human MM cell lines (HMCL).
Materials and methods
Cell culture
The HMCL L-363, JJN-3, KMS-12-BM, U-266 and AMO-1 were purchased from the “Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH” (DSMZ, Braunschweig, Germany), MM.1S from LGC Biolabs (Wesel, Germany) and KMS-11 was acquired from the Japanese Collection of Research Bioresources Cell Bank (JCRB1179).
All cell lines were cultured in RPMI-1640 medium (Thermo Fisher Scientific, Waltham, MA, USA) supplemented with 10% fetal bovine serum (FBS), 2 mM L-glutamine and 1 mM sodium pyruvate (all from PAN-Biotech, Aidenbach, Germany) at 37 °C and 5% CO2 for a maximum of 3 months and regularly tested for mycoplasma (VenorGEM One-Step kit (Minerva Biolabs, Berlin, Germany)). Cell lines were authenticated using the short tandem repeats profiling.
siRNA-mediated knockdown
HMCL were kept in culturing medium supplemented with 15% FBS for one night before transfection. 6 × 106 cells per condition were washed in PBS and resuspended in 200 µl unsupplemented RPMI-1640 medium containing 2.5 µM scrambled siRNA (scr-siRNA) (All Stars Negative Control siRNA, QIAGEN, Hilden, Germany, SI03650318) or ADAM8-, ADAM9- or ADAM15-specific siRNA (Supplementary Tables S1 and S2) immediately before electroporation in 2 mm cuvettes using the Gene Pulser Xcell electroporation system (BIO-RAD, Hercules, CA, USA) with an exponential program (capacitance: 980 µF) at 180 V (AMO-1, JJN-3, L-363), 200 V (MM.1S, KMS-11, KMS-12-BM) or 230 V (U-266) [26]. 200 µl unsupplemented medium were added to the cuvettes immediately after electroporation, the cells transferred into tubes containing further medium and incubated for 5 min before being transferred into 6-well plates containing 7 ml of prewarmed electroporation medium (EP medium) (RPMI supplemented with 15% FBS, 1% P/S, 2 mM L-glutamine). After 24 h, density gradient centrifugation was performed to separate living cells from dead cells. The cells were centrifuged, resuspended in 2.5 ml EP-medium mixed with 750 µl OptiPrep (Serumwerk, Bernburg, Germany)) and the suspension overlaid with 200 µl PBS. After centrifugation at 3500 rpm for 7 min, the viable cells were transferred from the medium-PBS interface into fresh EP-medium, centrifuged at 1000 rpm for 3 min, resuspended in a suitable amount of EP-medium and transferred to fresh 6-well plates to be incubated for another 24 h. 48 h after electroporation, cells were pelleted and lysates or RNA prepared for Western blotting or RNA sequencing, respectively. SiRNA knockdowns were performed at least three times in independent experiments for the assessment with Western blot. RNA was only extracted from one experiment.
Western blotting
Protein extraction, SDS-PAGE and Western blotting were performed as previously described [27]. 20 µg protein were loaded. Antibodies are listed in Supplementary Table S1. Band intensities were evaluated using the “gels” tool in Image J. Intensities for each marker were normalized to the corresponding GAPDH signal detected on the same blot. For siRNA knockdown experiments, the normalized expression of the siRNA samples was subsequently normalized to the respective scr-siRNA control sample. Western blot quantifications were statistically evaluated using the two-tailed t-test.
Patient samples
RNA sequencing of 73 previously CD138-sorted samples from 51 patients (“validation cohort”) was performed. For patient characteristics and corresponding experimental data see Supplementary Table S3. The study was approved by the Ethics Committee of the Medical Faculty, University of Würzburg (reference numbers 76/13 and 149/23-am). All methods were performed in accordance with the relevant guidelines and regulations. Informed consent was obtained from all subjects.
RNA sequencing
RNA extraction and sequencing
One RNA sequencing dataset was available from the Multiple Myeloma Research Foundation (MMRF CoMMpass study; 921 samples from 806 patients).
RNA from primary samples of the validation cohort was extracted using the DNA/RNA Micro kit (QIAGEN #80284) according to the manufacturers’ instructions. Following mRNA library preparation (Illumina), 100 bp paired end sequencing was performed on a NovaSeq (Illumina). Mapping was performed with STAR v2.7.2b and counts generated with featureCounts v1.6.4. Non-integer counts were rounded in R prior to analysis with DESeq2.
RNA from siRNA knockdown experiments was extracted from approximately 5 × 105 cells using the RNeasy mini kit (QIAGEN #74104), followed by Illumina mRNA library preparation. 150 bp paired end sequencing using a minimum of 200 ng total RNA were performed on a NovaSeq. Reads were mapped with Hisat2 v2.0.5 and counts generated with featureCounts v1.5.0-p3.
Sequencing quality parameters are summarized in Supplementary Table S4. Raw data has been uploaded to the European Genome-Phenome Archive (Accession: EGAS50000000392).
Analysis of differentially expressed genes
Differentially expressed genes between primary MM samples with the highest or lowest GE of ADAM8/9/15 (n = 92/condition in the MMRF cohort; n = 18/condition in the validation cohort) or before and after siRNA knockdown of ADAM8/9/15 in HMCL (n = 5–7/condition) were detected using DESeq2 in R [28]. For more details and information concerning sample grouping see supplementary methods. Principal component analysis (PCA) was performed on the vst-transformed DESeq datasets using the R packages DESeq2 [28], magrittr [29] and ggplot2 [30]. Volcano plots were created using EnhancedVolcano [31].
Gene set enrichment analysis (GSEA)
GSEA (GSEA 4.3.2, Broad Institute [32]) was performed on normalized counts matrices obtained from DESeq2 using hallmark gene sets (h.all.v2023.1.Hs.symbols.gmt) with a weighted enrichment statistic. FDR q values < 0.25 were considered significant.
Statistical comparison of ADAM8/9/15 gene expression between samples with different cytogenetic backgrounds and clinical parameters
ADAM8/9/15 GE (TPM) was compared between samples with or without high-risk cytogenetic abnormalities using the Mann–Whitney U test in both cohorts. P values were adjusted using the Benjamini–Hochberg correction in R.
In the MMRF cohort, ADAM8/9/15 GE (TPM) was additionally compared between the baseline sample and sample(s) taken from the same patient at progressive disease using the Wilcoxon-test (n = 59 patients). When more than one progressive disease sample was available for a patient, samples were treated as replicates.
In the validation cohort, ADAM8/9/15 GE (TPM) was compared between samples from patients with (n = 9) or without (n = 41) extramedullary disease (EMD) at the time of biopsy and between samples obtained from untreated patients (n = 13) and relapsed/refractory MM (RRMM) (n = 34) using the Mann–Whitney-U test. Where more than one sample was available from a patient, the mean ADAM8/9/15 GE of all samples was used if all samples belonged to the same group.
Immunohistochemical staining and analysis
MYC was previously stained on formalin-fixed paraffin-embedded (FFPE) bone marrow material from samples within the validation cohort using immunohistochemistry (IHC) [33]. Ki67 (MIB-1, Dako, 1:800) and CD138 (MI15, Dako, 1:100) were stained on consecutive slides after boiling in citric acid (pH 6.0).
The percentage of Ki67-positive CD138-positive tumor cells was evaluated by an expert hematopathologist (AR). Enrichment of Ki67high (≥30% Ki67+ CD138+ cells) or MYChigh (≥40% MYC+ CD138+ cells) samples in the ADAM8/9/15high/low (GE >/≤ mean) groups was investigated using Fisher’s exact test. MYC (% MYC+ CD138+ cells) and Ki67 expression (% Ki67+ CD138+ cells) was additionally compared between ADAM8/9/15high/low samples using the Mann–Whitney-U test.
Survival analyses
High GE of ADAM8, ADAM9 and ADAM15 was correlated with OS and PFS using the Kaplan-Meier method and log rank test in the validation cohort. Patients were grouped as ADAM8/9/15high when the median GE of all samples from the respective patient was higher than the mean GE (TPM) of all samples in the cohort.
Multivariate survival analysis was performed using the Cox proportional hazards model with high/low MYC, Ki67 and ADAM8/9/15 expression or high-risk cytogenetic abnormalities and ADAM8/9/15 expression as variables. Patients were MYChigh or Ki67high when at least one sample had ≥40% MYC-positive or ≥30% Ki67-positive MM cells (measured by IHC). A cytogenetic abnormality was considered to be present in a patient when it was detected in at least one sample. Analyses were performed using GraphPad Prism 9.
Results
High expression of ADAM8, ADAM9 and ADAM15 is associated with high-risk cytogenetic abnormalities
Looking at clinically relevant molecular parameters, we compared the GE of ADAM8/9/15 between patient samples with or without high-risk cytogenetic abnormalities (defined in ref. [1]) in two patient cohorts (MMRF, in-house validation). The 1q gain/amplification was associated with a significantly higher GE of ADAM8, ADAM9 and ADAM15 in the MMRF cohort (Table 1). Additionally, ADAM8 GE was significantly higher in samples with TP53 abnormalities or del17p in the MMRF cohort or validation cohort, respectively (Table 1). ADAM9 GE was higher in samples with the translocation t(14;16) in the MMRF cohort (Table 1). A significant but inconsistent association between ADAM8 and ADAM9 GE levels and the presence of t(4;14) was also observed in the MMRF cohort (Table 1).
Table 1.
Comparison of ADAM8, ADAM9 and ADAM15 (GE) (TPM) between samples with and without cytogenetic abnormalities associated with a high risk status (defined in ref. [1]).
| Cytogenetic abnormality absent vs. present | |||||||
|---|---|---|---|---|---|---|---|
| MMRF cohort | |||||||
| Abnormality | n = | ADAM8 GE | padj. | ADAM9 GE | padj. | ADAM15 GE | padj. |
| del17p | 428 vs. 58 | 2.78 vs. 3.82 | 0.105 | 4.95 vs. 6.50 | 0.285 | 10.69 vs. 9.841 | 0.687 |
| TP53 | 435 vs. 88 | 2.74 vs. 4.06 | 0.024 | 4.84 vs. 6.32 | 0.067 | 10.47 vs. 11.18 | 0.296 |
| 1q gain/amp | 352 vs. 222 | 2.72 vs. 3.49 | 0.024 | 4.21 vs. 6.69 | 1.31E−07 | 8.56 vs. 13.14 | 1.80E−13 |
| t(4;14) | 521 vs. 119 | 3.35 vs. 2.00 | 0.013 | 4.79 vs. 6.31 | 0.013 | 10.25 vs. 10.60 | 0.687 |
| t(14;16) | 539 vs. 57 | 3.02 vs. 3.82 | 0.285 | 4.84 vs. 6.78 | 0.031 | 10.08 vs. 12.00 | 0.077 |
| t(14;20) | 357 vs. 10 | 2.53 vs. 3.10 | 0.767 | 4.93 vs. 6.13 | 0.785 | 10.21 vs. 11.15 | 0.767 |
| Validation cohort | |||||||
| Abnormality | n = | ADAM8 GE | padj. | ADAM9 GE | padj. | ADAM15 GE | padj. |
| del17p | 43 vs. 18 | 2.22 vs. 7.01 | 0.015 | 2.69 vs. 3.38 | 0.477 | 8.67 vs. 11.84 | 0.463 |
| TP53_mut | 50 vs. 10 | 2.93 vs. 3.99 | 0.563 | 2.35 vs. 4.82 | 0.170 | 9.83 vs. 14.88 | 0.284 |
| 1q gain/amp | 34 vs. 21 | 2.75 vs. 3.49 | 0.765 | 2.41 vs. 3.29 | 0.765 | 10.03 vs. 7.15 | 0.912 |
| t(4;14) | 42 vs. 18 | 2.67 vs. 4.33 | 0.463 | 2.69 vs. 1.90 | 0.981 | 9.83 vs. 7.09 | 0.942 |
| t(14;16) | 54 vs. 2 | 2.83 vs. 6.41 | 0.563 | 2.67 vs. 5.28 | 0.563 | 9.19 vs. 49.17 | 0.284 |
Statistical test was Mann–Whitney-U. P values were adjusted (padj.) for testing of multiple hypotheses using the Benjamini–Hochberg procedure in R. Translocation status for t(14;20) was not available in the validation cohort. Amp: amplification.
In summary, supporting a clinical relevance, the presence of at least one high-risk cytogenetic abnormality was associated with a higher GE of ADAM8/9/15 in the MMRF and/or validation cohort.
Adding to our previous findings, where a high GE of ADAM8, ADAM9 and ADAM15 was associated with shorter PFS and OS in the MMRF cohort [6], multivariate survival analyses considering ADAM8/9/15 GE and the respective cytogenetic abnormalities which were associated with a high ADAM8/9/15 GE (Table 1) revealed high ADAM8/9/15 GE as independent prognostic markers for shorter PFS and OS in the MMRF cohort (Supplementary Table S5).
Furthermore, high GE of ADAM8, ADAM9 and ADAM15 was also associated with a significantly shorter survival in the newly sequenced validation cohort (Supplementary Fig. S1) and multivariate survival analysis verified high ADAM8 GE as prognostic for shorter OS and PFS independent of the presence of del17p (Supplementary Table S6).
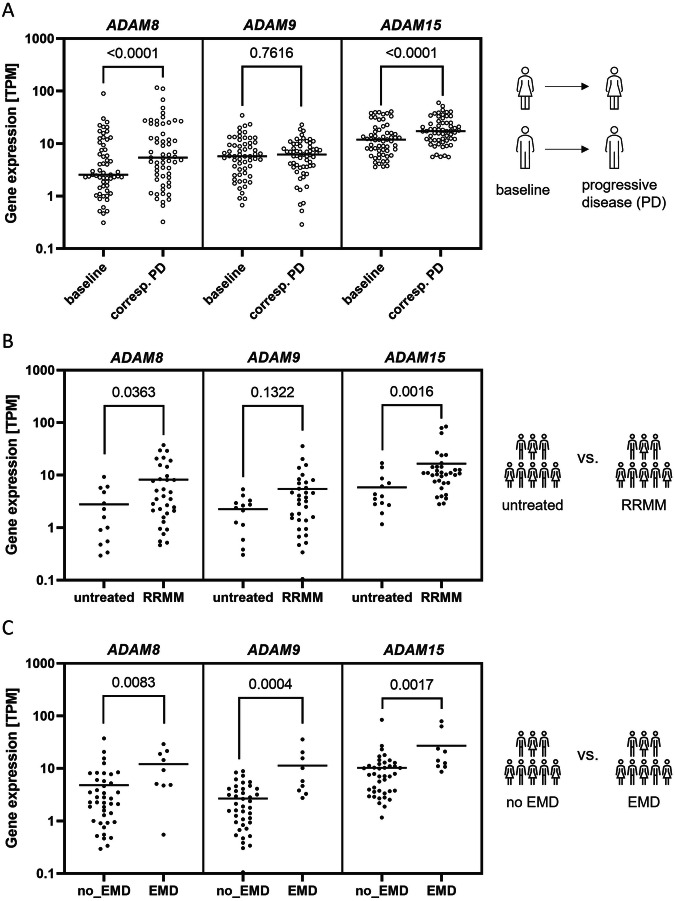
ADAM8, ADAM9 and ADAM15 upregulation is associated with progressive disease in MM
Next, we assessed the role of ADAM8, ADAM9 and ADAM15 GE in the context of MM progression in both patient cohorts. Analysis of paired samples revealed a significant upregulation of ADAM8 and ADAM15 GE in samples obtained at a stage of progressive disease compared to the corresponding baseline sample collected from the same patient in the MMRF cohort (Fig. 1A). ADAM8 and ADAM15 GE was also significantly higher in RRMM samples compared to samples from untreated patients in the validation cohort (Fig. 1B). Additionally, MM samples from patients with EMD at the time of biopsy had a significantly higher ADAM8, ADAM9 and ADAM15 GE than samples from patients with no EMD in the validation cohort (Fig. 1C).
Fig. 1. High expression of ADAM8, ADAM9 and ADAM15 is associated with progressive disease.
A Comparison of ADAM8, ADAM9 and ADAM15 GE between the baseline sample (first sample acquired when patient entered the study) and corresponding samples taken from the same patient at a stage of progressive disease (corresp. PD) in the MMRF cohort (n = 59 patients in analysis). Samples were treated as replicates if more than one PD sample was available for a patient. Statistical test was Wilcoxon-test. Lines show the mean GE. B Comparison of ADAM8, ADAM9 and ADAM15 GE between unpaired samples (except for 2) obtained from untreated patients (n = 13) and RRMM (n = 34) from the validation cohort. When more than one sample taken at the same stage was available for a patient, the mean was used. Statistical test was Mann–Whitney-U. Lines show the mean GE. C Comparison of ADAM8, ADAM9 and ADAM15 GE between unpaired samples from patients with or without extramedullary disease (EMD: n = 9 or no_EMD: n = 41) at biopsy in the validation cohort. Where more than one sample with the same EMD status was available from one patient, the mean GE of these samples was used. One patient acquired EMD within the course of the study, the remaining patients did not change groups. Statistical test was Mann–Whitney-U. Lines show the mean GE. Patient information and treatment for the validation cohort is summarized in Supplementary Table S3.
In summary, high GE of ADAM8, ADAM9 and ADAM15 was verified as prognostic for shorter patient survival and associated with high-risk cytogenetics as well as disease progression.
ADAM8/9/15 influence proliferation and survival signaling in MM
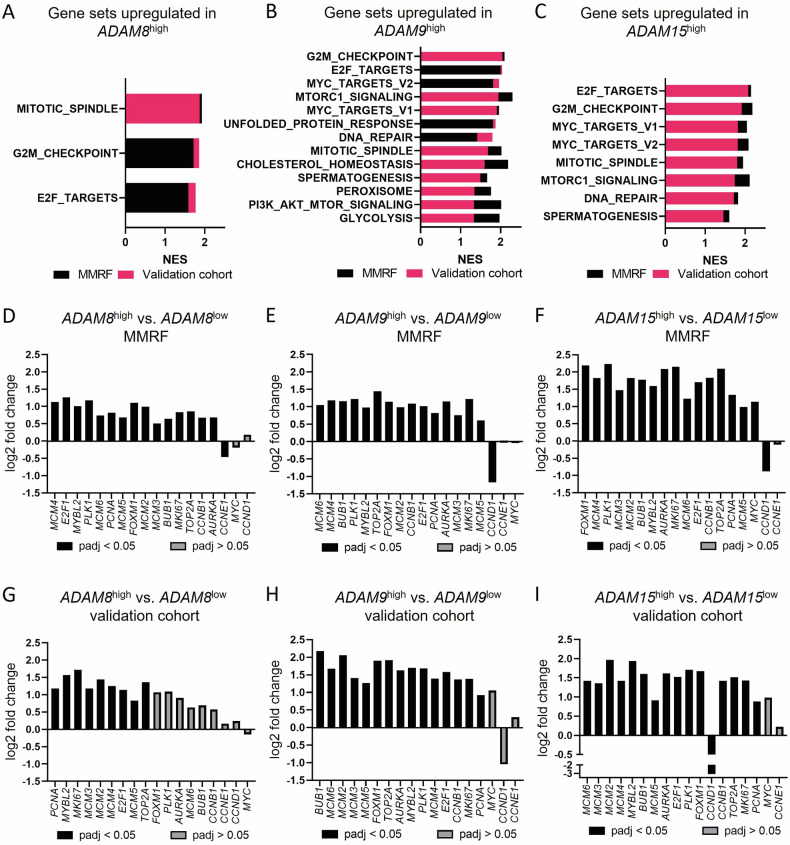
To gain insight into which signaling pathways may be regulated by ADAM8/9/15 in MM, we compared the GE profiles of ADAM8/9/15high and ADAM8/9/15low patient samples from both the MMRF and validation cohort using GSEA. For PCA and volcano plots of RNA sequencing data see Supplementary Figs. S2–S4. Differentially expressed genes are summarized in Supplementary Table S7. There was a considerable overlap of significantly enriched gene sets (FDR q value < 0.25) between the two patient cohorts and between the analyses for the different ADAM genes (Fig. 2A–C). The vast majority of these gene sets were associated with cell cycle, proliferation and survival signaling (Fig. 2A–C). More precisely, the gene sets “G2/M checkpoint”, “E2F targets” and “mitotic spindle” were significantly enriched in ADAM8/9/15high samples from both patient cohorts (Fig. 2A–C) and “DNA repair”, “MYC targets” and “MTORC1 signaling” were enriched in ADAM9/15high samples from both cohorts (Fig. 2B, C). For a summary of all enriched gene sets in the individual cohorts see Supplementary Table S8.
Fig. 2. ADAM8/9/15 expression levels influence proliferation signaling in MM.
Summary of gene sets where a significant enrichment (FDR q value < 0.25) was found in A ADAM8high, B ADAM9high or C ADAM15high patient samples from both the MMRF (black) and validation cohort (red). NES Normalized enrichment score. A summary of all enriched gene sets is shown in Supplementary Table S8. D–I Log2 fold change of expression of commonly used proliferation markers between ADAM8/9/15high vs. ADAM8/9/15low primary MM samples. Top 10% (MMRF (D–F)) or 25% (validation cohort (G–I)) of samples with the highest/lowest ADAM8/9/15 GE were included. padj values (p value adjusted for multiple hypothesis testing) increase from left to right. Significantly differentially expressed genes (padj < 0.05) have black bars, genes with padj > 0.05 are depicted in gray. A summary of all differentially expressed genes is shown in Supplementary Table S7.
We subsequently assessed the differential expression of common proliferation markers [34, 35] between ADAM8/9/15high and ADAM8/9/15low patient samples. Virtually all of the significantly differentially expressed (padj<0.05) proliferation markers investigated were upregulated in the ADAM8/9/15high samples from both patient cohorts (Fig. 2D–I). The only exceptions were the significant downregulation of Cyclin E1 (CCNE1) in ADAM8high samples from the MMRF cohort and of Cyclin D1 (CCND1) in ADAM9high samples from the MMRF cohort and ADAM15high samples from both cohorts (Fig. 2D, E, F, I).
Since CCND1 upregulation can be caused by the translocation t(11;14) [36], we assessed whether there was an enrichment of samples with t(11;14) in the ADAM9/15low samples. Samples with t(11;14) were significantly enriched in the ADAM9low samples from the MMRF but not the ADAM15low samples from either of the cohorts (Supplementary Fig. S5).
In summary, high ADAM8/9/15 expression levels influenced important proliferation and survival signaling pathways in MM patients from two different cohorts.
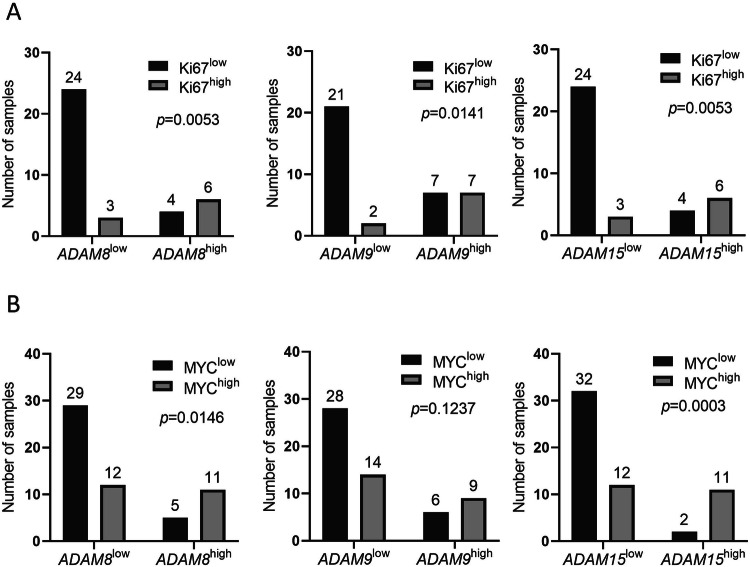
High ADAM8/9/15 expression correlates with high Ki67 and MYC protein expression
Since the high expression of ADAM8/9/15 was associated with an upregulation of proliferation marker gene expression (Fig. 2), we subsequently assessed Ki67 and MYC protein expression detected by IHC in ADAM8/9/15high and ADAM8/9/15low patient samples from the validation cohort (Fig. 3, Supplementary Figs. S6, S7). We found a significant association between high ADAM8/9/15 and high Ki67 expression (Fig. 3A, Supplementary Fig. S6). Moreover, high ADAM8 and ADAM15 expression were associated with high MYC expression levels (Fig. 3B, Supplementary Fig. S6).
Fig. 3. Ki67 and MYC protein expression in samples from the validation cohort.
A Samples with high Ki67 protein expression determined by IHC are significantly enriched in ADAM8/9/15high patient samples from the validation cohort. B Samples with high MYC protein expression determined by IHC are significantly enriched in ADAM8/15high but not in ADAM9high patient samples from the validation cohort. ADAM8/9/15high/low: ADAM8/9/15 GE>/≤ mean of all samples. Ki67high/low: ≥30%/<30% Ki+ CD138+ cells. MYChigh/low: ≥40%/<40% MYC+ CD138+ cells. Statistical test was Fisher’s exact test. For exemplary Ki67 stainings see Supplementary Fig. S7. For MYC stainings see ref. [33].
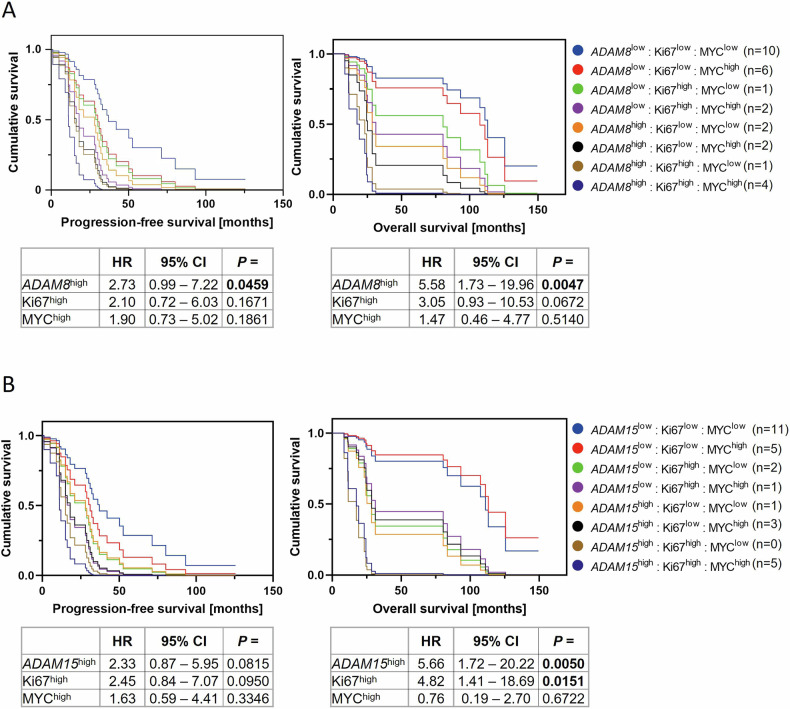
Since high Ki67 and MYC protein expression have been shown to correlate with shorter PFS and/or OS in MM [33, 37–39], the prognostic value of high ADAM8/9/15 GE was subsequently reassessed in multivariate survival analyses. Cox regression considering Ki67 and MYC protein and ADAM8 GE confirmed high ADAM8 GE as an independent prognostic marker for shorter PFS and OS (Fig. 4A).
Fig. 4. High ADAM8 and ADAM15 expression levels are independent prognostic markers in MM.
Cox proportional hazards model assessing the effect of high Ki67, MYC and (A) ADAM8 or (B) ADAM15 expression on progression-free survival (left) and overall survival (right) in the validation cohort. ADAM8/15high patients have an ADAM8/15 GE >mean of all samples. Ki67high patients had at least one sample with ≥30% Ki67-expressing CD138+ cells. MYChigh was assigned to a patient if at least one sample contained ≥40% MYC-expressing MM cells. n = 28 patients. HR hazard ratio, CI confidence interval.
Cox regression considering both ADAM9 and Ki67 expression only verified high Ki67 expression as an independent prognostic marker for shorter PFS and OS (Supplementary Fig. S8).
Multivariate survival analyses considering ADAM15, Ki67 and MYC expression confirmed both the high ADAM15 and Ki67 expression as independent predictors for worse OS but not for PFS (Fig. 4B). For Cox regressions considering only ADAM8/15 GE and either Ki67 or MYC expression see Supplementary Figs. S9, S10.
siRNA knockdown of ADAM8/9/15 influences proliferation/survival signaling in HMCL
In an experimental approach using siRNA knockdowns of ADAM8, ADAM9 or ADAM15 in HMCL, we aimed to verify the influence of ADAM8/9/15 expression levels on the signaling pathways enriched in the GSEA comparing the expression profiles of ADAM8/9/15high/low patient samples.
ADAM8/9/15 protein expression was assessed prior to siRNA knockdowns in seven HMCL by Western blotting. ADAM8 and ADAM15 were expressed in 5/7 HMCL and ADAM9 was expressed in all seven HMCL (Supplementary Fig. S11). Knockdowns were only performed in the HMCL with considerable expression of the respective ADAMs.
Similar to what was observed in the two patient cohorts, GSEA comparing the gene expression profiles of HMCL before (scr-siRNA control) and after siRNA knockdown of ADAM8/9/15 revealed a significant enrichment of gene sets associated with proliferation, cell cycle and survival (Supplementary Fig. S12, Supplementary Table S8).
Moreover, we assessed the effect of ADAM8/9/15 siRNA knockdown on further signaling pathways of known importance in MM [39–42] which were highly enriched in the GSEA of patient samples using Western blots.
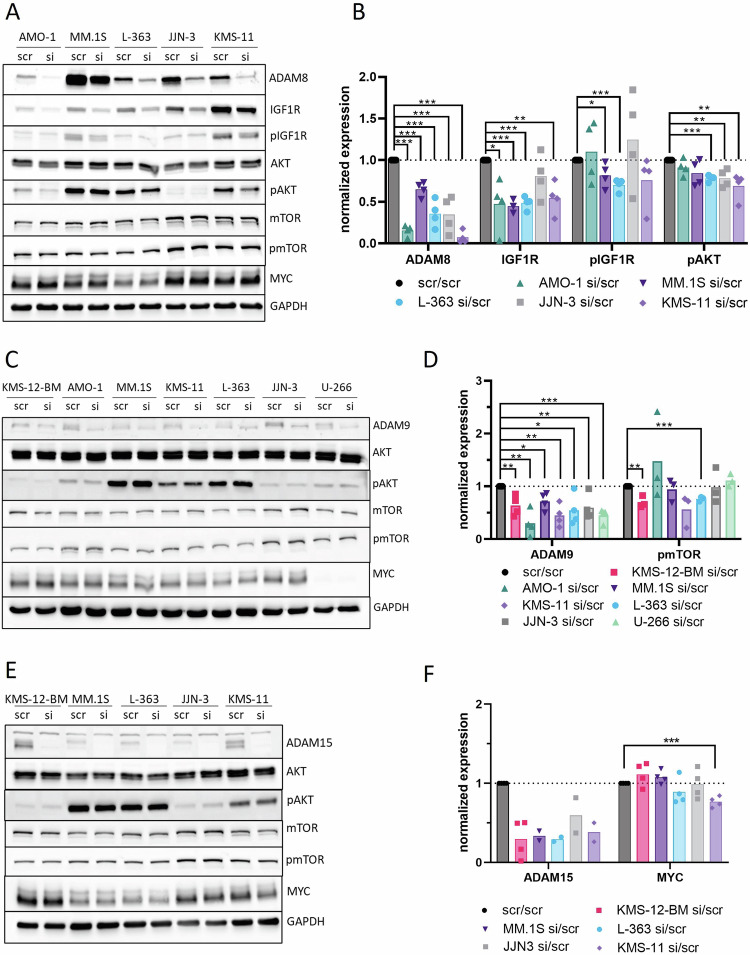
“PI3K/AKT/MTOR signaling” was the top enriched gene set (highest normalized enrichment score) in the ADAM8high patient samples from the MMRF cohort (Supplementary Table S8). ADAM8 knockdown significantly reduced the expression of the receptor tyrosine kinase insulin-like growth factor 1 receptor (IGF1R), which is known to regulate this pathway [43], by approximately half in 4/5 HMCL (Fig. 5A, B). Moreover, pIGF1R levels were clearly reduced in 3/5 HMCL (Fig. 5A, B). AKT expression was unaffected while pAKT was significantly reduced in 3/5 HMCL (Fig. 5A, B). A conclusive effect on mTOR expression or activation was not observed (Fig. 5A, B).
Fig. 5. ADAM8/9/15 siRNA knockdowns in HMCL.
A–F Evaluation of expression and activation of members of the PI3K/AKT/mTOR signaling pathway and MYC expression using Western blotting after ADAM8/9/15 siRNA knockdown. Cells were transfected with either scr-siRNA control (scr) or ADAM8/9/15-specific siRNA (si) in four independent rounds of experiments, respectively. A, C, E Representative Western blots. Pan and phospho-markers were detected on separate blots and only the relevant area of the blots is shown. Housekeeper (GAPDH) was detected on each blot, representative GAPDH staining is shown. B, D, F Summary of normalized expression for all evaluable rounds of experiments for markers where an effect was observed. The expression of each marker was first normalized to the expression of the housekeeper and subsequently the siRNA samples were normalized to the scr-siRNA controls. Each data point represents one independent round of experiments. Bars show the mean. Statistical test was two-tailed t-test. * for p < 0.05, ** for p < 0.01, *** for p < 0.001. B n = 4 for all markers statistically evaluated except for IGF1R in MM.1S (n = 3). D n = 4 for ADAM9, n = 3 for pmTOR. F n = 4 for MYC. ADAM15 was only evaluable for all cell lines in two rounds of experiments because of a complete lack of bands for the siRNA transfected HMCLs in the other rounds due to a complete knockdown (see blot in E). The difference in ADAM15 expression levels between scr and si was therefore not statistically evaluated.
“MTORC1 signaling” was the top enriched gene set in ADAM9high patient samples from the MMRF cohort (Supplementary Table S8) and “MYC targets” were enriched in ADAM9high patient samples from both cohorts as well as in the comparison of HMCL before and after ADAM9 siRNA knockdown (Fig. 2B, Supplementary Fig. S12). AKT, pAKT, mTOR or MYC expression were not affected by ADAM9 siRNA knockdown on protein level. However, pmTOR was significantly reduced in KMS-12-BM and L-363 and also clearly reduced by 44% in KMS-11 (Fig. 5C, D).
The gene set “MTORC1 signaling” was enriched in ADAM15high samples from both cohorts and in the comparison of HMCL before and after ADAM15 siRNA knockdown (Fig. 2C, Supplementary Fig. S12). “MYC targets” were enriched in ADAM15high samples from both patient cohorts (Fig. 2C). A conclusive effect of ADAM15 siRNA knockdown on the mTOR signaling members assessed herein was not observed in Western blots. However, MYC was significantly downregulated in KMS-11 (Fig. 5E, F).
In summary, similar to our observations in the two patient cohorts, ADAM8/9/15 expression levels influenced survival and proliferation signaling pathways in an experimental approach using siRNA knockdowns in HMCL.
Discussion
Our study validates the previously proposed [6] prognostic potential of a high ADAM8, ADAM9 and ADAM15 GE in a newly sequenced patient cohort and investigates the thus far scarcely studied clinical and functional role of ADAM8, ADAM9 and ADAM15 in two MM patient cohorts (MMRF CoMMpass study cohort and our own validation cohort) and seven HMCL.
Focusing on clinical aspects, we found an association between the presence of high-risk cytogenetic abnormalities and an increased ADAM8/9/15 GE. Remarkably, the 1q amplification/gain was associated with a higher GE of all three ADAMs assessed herein. This association was particularly strong for ADAM15, which is in line with the fact that ADAM15 is encoded on chromosome 1q21.3. Since high-risk cytogenetic abnormalities are associated with shorter patient survival and e.g. the incidence of 1q amplifications/gains increases with MM progression [44–47], the role of ADAM8/9/15 in the prognosis and progression of MM was investigated in more detail.
Underlining the clinical relevance of ADAM8/9/15, multivariate survival analyses verified high ADAM8/9/15 expression levels as robust prognostic markers for shorter patient survival independent of the association with high-risk cytogenetics in this study.
Apart from high-risk cytogenetic abnormalities, high ADAM8 and ADAM15 expression also correlated with high MYC protein expression and high ADAM8, ADAM9 and ADAM15 expression correlated with high Ki67 protein expression in MM patients in the current study. High Ki67 expression is a known prognostic marker in MM [37, 38]. Moreover, MM is generally considered to be MYC-driven [41] and high MYC expression is known to affect OS [33, 39] and to correlate with a high proliferation index (Ki67) [39]. Nevertheless, high expression levels of ADAM8 and ADAM15 were also verified as prognostic markers independent of Ki67 and MYC expression levels in multivariate survival analyses, underlining their potential suitability as biomarkers.
Next, we found an upregulation of ADAM8, ADAM9 and ADAM15 in patients with EMD, which is associated with aggressive and progressive disease [48], in the validation cohort. Similarly, ADAM8 and ADAM15 GE also increased with disease progression in the MMRF cohort and ADAM8 and ADAM15 GE increased in RRMM compared to untreated samples from the validation cohort. These results imply a possible role for ADAM8/9/15 in MM progression, relapse and therapy resistance. Accordingly, expression levels of ADAM8, ADAM9 and ADAM15 have all been shown to be involved in the progression and/or metastasis/invasion of various solid cancers [9, 11–13, 15–18, 20, 21]. Furthermore, ADAM8 has been linked to chemoresistance in solid cancers [9] and tyrosine kinase inhibitor therapy resistance in chronic myeloid leukemia cells [49].
GSEA comparing ADAM8/9/15high/low MM patient samples of two different cohorts or HMCL before and after ADAM8/9/15 siRNA knockdown revealed an upregulation of gene sets associated with proliferation and cell cycle/growth in the ADAM8/9/15high groups, with “G2/M checkpoint”, “E2F targets”, “MYC targets” and “MTORC1 signaling” among the most frequently upregulated gene sets. The considerable overlap of gene sets that were enriched when comparing ADAM8/9/15high/low patient samples from the two different cohorts and when comparing HMCL before and after active downregulation of ADAM8/9/15 by siRNA knockdown supports the reliability of these results.
Interestingly, it has been shown that triple-relapsed MM compared to newly diagnosed or relapsed samples pre-daratumumab exposure [50], and extramedullary MM compared to newly diagnosed MM [51] upregulate E2F and MYC targets as well as the G2/M checkpoint gene set, further underscoring the association between ADAM8/9/15 upregulation and disease progression.
Consistent with the GSEA results, which suggested an influence of ADAM8/9/15 expression on proliferation signaling, almost all proliferation markers [34, 35] were upregulated in ADAM8/9/15high samples on RNA level in both patient cohorts. The only exceptions were the downregulation of CCND1 in ADAM9/15high, CCNE1 in the ADAM8high samples in the MMRF and/or the validation cohort. CCND1 is a known player in MM due to the common translocation t(11;14) [36]. The enrichment of cases with t(11;14) in the ADAM9low samples from the MMRF cohort might explain why CCND1 appeared to be downregulated in the ADAM9high samples. However, samples with t(11;14) were evenly distributed between ADAM15high/low samples from both cohorts, and the downregulation of CCND1 is therefore most likely explained by the cyclic up- and downregulation of cyclins depending on the cell cycle phase [52]. In line with the enrichment of the G2/M checkpoint gene set, CCNB1, important for the G2/M transition [52], was upregulated where CCND1 or CCNE1, important factors in G1 and S phase [52], were downregulated.
MKI67 (encoding Ki67), one of the most well-known proliferation markers, which is used in routine tumor diagnostics to assess the proliferation index, was significantly upregulated in ADAM8/9/15high samples from both cohorts on RNA level and high ADAM8/9/15 expression also correlated with high Ki67 protein expression, further strengthening the hypothesis that high ADAM8, ADAM9 and ADAM15 expression may be associated with increased proliferation.
In line with that, this study found evidence that ADAM8 might influence the PI3K/AKT/mTOR signaling pathway, as this pathway was enriched the most in ADAM8high patient samples and IGF1R expression and activation as well as AKT activation was downregulated by ADAM8 siRNA knockdown in HMCL. The IGF1R/PI3K/AKT/mTOR signaling cascade is commonly activated in MM and regulates MM cell growth, proliferation and survival [40, 53–55]. Furthermore, an influence of ADAM8 expression on MAPK and AKT signaling has also been observed in other cancers [11, 12], underscoring our current findings in MM.
Genes associated with MTORC1 signaling showed a high enrichment in ADAM9/15high compared to ADAM9/15low patient samples. Studies have described mTOR as an important factor in MM, influencing e.g. proliferation, growth, survival, invasion and chemoresistance [40]. In line with the GSEA results, ADAM9 siRNA knockdown reduced mTOR activation in several HMCL. While pAKT was largely unaffected, this could be explained by alternative kinases phosphorylating mTOR [56]. No conclusive effect on the members of the MTORC1 signaling pathway assessed herein was observed upon ADAM15 siRNA knockdown. However, since MTORC1 activity can be influenced by a number of factors (phosphorylation by various kinases, interacting partners in the complex, nutrient supply, localization [57]), conclusively verifying the effect of ADAM9/15 expression on MTORC1 signaling is beyond the scope of this project. Nevertheless, these data still provide further evidence that ADAM9 may influence MTORC1 signaling in MM, as has been described for colorectal cancer cells [58].
Consistent with the influence on proliferation signaling in MM observed herein, ADAM8, 9 and 15 have been shown to influence the proliferation of e.g. hepatocellular, renal and prostate cancer [12, 13, 19, 22].
Considering the various functions of ADAMs, possible mechanisms by which ADAM8/9/15 expression levels might influence proliferation signaling could be e.g. the cleavage of growth factors and receptors or the interaction with integrins [7–9]. For instance, it has been shown that ADAM9, expressed on MM cells, can interact with integrin αvβ5 on osteoblasts and thereby promote their production of interleukin-6 [24] which, in turn, can stimulate the proliferation of MM cells and prevent apoptosis [59]. Nevertheless, the exact mechanisms by which ADAM8/9/15 influence proliferation signaling in MM remain to be elucidated in future studies.
In conclusion, this study showed that high expression levels of ADAM8, ADAM9 and ADAM15 are interesting biomarkers of prognostic relevance in MM, that are linked to a high-risk status and disease progression and influence several important survival and proliferation signaling pathways. Further studies should therefore assess the potential usefulness of targeting these ADAMs therapeutically.
Supplementary information
Supplementary methods, figures and tables S1, S5, S6
Acknowledgements
This work was supported by the Deutsche Krebshilfe (EL: project number 70112693) and the Open Access Publication Fund of the University of Würzburg. RE, FJ and T.Ste were supported by the Deutsche Forschungsgemeinschaft (DFG) (RE: SPP microBONE grant EB 447/10-1 (project number 491715122); FJ: grant JU426/6-1 (project number 370022528) and JU 426/10-1 (project number 491715122); T.Ste (project number 442740310)). The MMRF datasets analyzed during the current study are available at https://research.themmrf.org and www.themmrf.org. A part of these data were generated as part of the Multiple Myeloma Research Foundation Personalized Medicine Initiatives (https://research.themmrf.org and www.themmrf.org).
Author contributions
EL and ME conceived and designed the research study; ME performed experiments; ME, MS, T.Ste and EL acquired data; EL supervised the study; ME, MR, AR and EL analyzed the data; ME, TStü, FJ, RE, TNH, RCB, AR and EL interpreted the data; ME drafted the manuscript and EL helped with writing and finalizing the manuscript; All authors revised the manuscript, approved the final version and will ensure that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Data availability
The datasets generated and/or analyzed during the current study have been uploaded to the European Genome-Phenome Archive (EGA) (Accession: EGAS50000000392).
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
The online version contains supplementary material available at 10.1038/s41408-024-01133-4.
References
- 1.Rajkumar SV. Multiple myeloma: 2022 update on diagnosis, risk stratification, and management. Am J Hematol. 2022;97:1086–107. 10.1002/ajh.26590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar SK, Dispenzieri A, Lacy MQ, Gertz MA, Buadi FK, Pandey S, et al. Continued improvement in survival in multiple myeloma: changes in early mortality and outcomes in older patients. Leukemia. 2014;28:1122–8. 10.1038/leu.2013.313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Durie BGM, Hoering A, Abidi MH, Rajkumar SV, Epstein J, Kahanic SP, et al. Bortezomib with lenalidomide and dexamethasone versus lenalidomide and dexamethasone alone in patients with newly diagnosed myeloma without intent for immediate autologous stem-cell transplant (SWOG S0777): a randomised, open-label, phase 3 trial. Lancet. 2017;389:519–27. 10.1016/S0140-6736(16)31594-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kawano Y, Moschetta M, Manier S, Glavey S, Görgün GT, Roccaro AM, et al. Targeting the bone marrow microenvironment in multiple myeloma. Immunol Rev. 2015;263:160–72. 10.1111/imr.12233 [DOI] [PubMed] [Google Scholar]
- 5.Karamanos NK, Theocharis AD, Piperigkou Z, Manou D, Passi A, Skandalis SS, et al. A guide to the composition and functions of the extracellular matrix. FEBS J. 2021;288:6850–912. 10.1111/febs.15776 [DOI] [PubMed] [Google Scholar]
- 6.Evers M, Schreder M, Stühmer T, Jundt F, Ebert R, Hartmann TN, et al. Prognostic value of extracellular matrix gene mutations and expression in multiple myeloma. Blood Cancer J. 2023;13:43. 10.1038/s41408-023-00817-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edwards DR, Handsley MM, Pennington CJ. The ADAM metalloproteinases. Mol Asp Med. 2008;29:258–89. 10.1016/j.mam.2008.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rocks N, Paulissen G, Hour ME, Quesada F, Crahay C, Guéders MM, et al. Emerging roles of ADAM and ADAMTS metalloproteinases in cancer. Biochimie. 2008;90:369–79. 10.1016/j.biochi.2007.08.008 [DOI] [PubMed] [Google Scholar]
- 9.Conrad C, Benzel J, Dorzweiler K, Cook L, Schlomann U, Zarbock A, et al. ADAM8 in invasive cancers: links to tumor progression, metastasis, and chemoresistance. Clin Sci. 2019;133:83–99. 10.1042/CS20180906 [DOI] [PubMed] [Google Scholar]
- 10.Duffy MJ, McKiernan E, O’Donovan N, McGowan PM. Role of ADAMs in Cancer Formation and Progression. Clin Cancer Res. 2009;15:1140–4. 10.1158/1078-0432.CCR-08-1585 [DOI] [PubMed] [Google Scholar]
- 11.Schlomann U, Koller G, Conrad C, Ferdous T, Golfi P, Garcia AM, et al. ADAM8 as a drug target in pancreatic cancer. Nat Commun. 2015;6:6175. 10.1038/ncomms7175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Awan T, Babendreyer A, Mahmood Alvi A, Düsterhöft S, Lambertz D, Bartsch JW, et al. Expression levels of the metalloproteinase ADAM8 critically regulate proliferation, migration and malignant signalling events in hepatoma cells. J Cell Mol Med. 2021;25:1982–99. 10.1111/jcmm.16015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qu H, Mao M, Wang K, Mu Z, Hu B. Knockdown of ADAM8 inhibits the proliferation, migration, invasion, and tumorigenesis of renal clear cell carcinoma cells to enhance the immunotherapy efficacy. Transl Res. 2024;266:32–48. 10.1016/j.trsl.2023.11.003 [DOI] [PubMed] [Google Scholar]
- 14.Grützmann R, Lüttges J, Sipos B, Ammerpohl O, Dobrowolski F, Alldinger I, et al. ADAM9 expression in pancreatic cancer is associated with tumour type and is a prognostic factor in ductal adenocarcinoma. Br J Cancer. 2004;90:1053–8. 10.1038/sj.bjc.6601645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shintani Y, Higashiyama S, Ohta M, Hirabayashi H, Yamamoto S, Yoshimasu T, et al. Overexpression of ADAM9 in non-small cell lung cancer correlates with brain metastasis. J. Cancer Res. 2004;64:4190–6. 10.1158/0008-5472.CAN-03-3235 [DOI] [PubMed] [Google Scholar]
- 16.Mazzocca A, Coppari R, De Franco R, Cho J-Y, Libermann TA, Pinzani M, et al. A Secreted Form of ADAM9 Promotes Carcinoma Invasion through Tumor-Stromal Interactions. Cancer Res. 2005;65:4728–38. 10.1158/0008-5472.CAN-04-4449 [DOI] [PubMed] [Google Scholar]
- 17.Zhou R, Cho WCS, Ma V, Cheuk W, So YK, Wong SCC, et al. ADAM9 Mediates Triple-Negative Breast Cancer Progression via AKT/NF-κB Pathway. Front Med. 2020;7:214. 10.3389/fmed.2020.00214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J, Ji Z, Qiao C, Qi Y, Shi W. Overexpression of ADAM9 Promotes Colon Cancer Cells Invasion. J Investigative Surg. 2013;26:127–33. 10.3109/08941939.2012.728682 [DOI] [PubMed] [Google Scholar]
- 19.Liu CM, Hsieh CL, He YC, Lo SJ, Liang JA, Hsieh TF, et al. In vivo targeting of ADAM9 gene expression using lentivirus-delivered shRNA suppresses prostate cancer growth by regulating REG4 dependent cell cycle progression. PloS One. 2013;8:e53795. 10.1371/journal.pone.0053795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dong DD, Zhou H, Li G. ADAM15 targets MMP9 activity to promote lung cancer cell invasion. Oncol Rep. 2015;34:2451–60. 10.3892/or.2015.4203 [DOI] [PubMed] [Google Scholar]
- 21.Lorenzatti Hiles G, Bucheit A, Rubin JR, Hayward A, Cates AL, Day KC, et al. ADAM15 Is Functionally Associated with the Metastatic Progression of Human Bladder Cancer. PloS One. 2016;11:e0150138. 10.1371/journal.pone.0150138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xu JH, Guan YJ, Zhang YC, Qiu ZD, Zhou Y, Chen C, et al. ADAM15 correlates with prognosis, immune infiltration and apoptosis in hepatocellular carcinoma. Aging. 2021;13:20395–417. 10.18632/aging.203425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoelzinger DB, Quinton SJ, Walters DK, Tschumper RC, Jelinek DF. Proteomic and Biological Analysis of Myeloma Cell Derived Extracellular Vesicles. Blood. 2018;132:5605. 10.1182/blood-2018-99-116926 [DOI] [Google Scholar]
- 24.Karadag A, Zhou M, Croucher PI. ADAM-9 (MDC-9/meltrin-γ), a member of the adisintegrin and metalloproteinase family, regulates myeloma-cell–induced interleukin-6 production in osteoblasts by direct interaction with the αvβ5 integrin. Blood. 2006;107:3271–8. 10.1182/blood-2005-09-3830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bret C, Hose D, Reme T, Kassambara A, Seckinger A, Meißner T, et al. Gene expression profile of ADAMs and ADAMTSs metalloproteinases in normal and malignant plasma cells and in the bone marrow environment. Exp Hematol. 2011;39:546–57.e8. 10.1016/j.exphem.2011.02.002 [DOI] [PubMed] [Google Scholar]
- 26.Steinbrunn T, Chatterjee M, Bargou RC, Stühmer T. Efficient Transient Transfection of Human Multiple Myeloma Cells by Electroporation – An Appraisal. PloS One. 2014;9:e97443. 10.1371/journal.pone.0097443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Weißbach S, Heredia-Guerrero SC, Barnsteiner S, Großhans L, Bodem J, Starz H, et al. Exon-4 Mutations in KRAS Affect MEK/ERK and PI3K/AKT Signaling in Human Multiple Myeloma Cell Lines. Cancers. 2020;12:455. 10.3390/cancers12020455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bache S, Wickham H. magrittr: A Forward-Pipe Operator for R. R package version 2.0.3. 2022. https://CRAN.Rproject.org/package=magrittr
- 30.Wickham H, Chang W, Wickham. MHJCedvutgogV. Package ‘ggplot2’. 2016;2:1–189. [Google Scholar]
- 31.Blighe K, Rana S, Lewis MJE. EnhancedVolcano: Publication-ready volcano plots with enhanced colouring and labeling. https://github.com/kevinblighe 2018.
- 32.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. J Proc Natl Acad Sci. 2005;102:15545–50. 10.1073/pnas.0506580102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leich E, Schreder M, Pischimarov J, Stühmer T, Steinbrunn T, Rudelius M, et al. Novel molecular subgroups within the context of receptor tyrosine kinase and adhesion signalling in multiple myeloma. Blood Cancer J. 2021;11:51. 10.1038/s41408-021-00442-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Iatropoulos MJ, Williams GM. Proliferation markers. Exp Toxicol Pathol. 1996;48:175–81. 10.1016/S0940-2993(96)80039-X [DOI] [PubMed] [Google Scholar]
- 35.Whitfield ML, George LK, Grant GD, Perou CM. Common markers of proliferation. Nat Rev Cancer. 2006;6:99–106. 10.1038/nrc1802 [DOI] [PubMed] [Google Scholar]
- 36.Bal S, Kumar SK, Fonseca R, Gay F, Hungria VT, Dogan A, et al. Multiple myeloma with t(11;14): unique biology and evolving landscape. Am J cancer Res. 2022;12:2950–65. [PMC free article] [PubMed] [Google Scholar]
- 37.Alexandrakis MG, Passam FH, Kyriakou DS, Dambaki K, Niniraki M, Stathopoulos E. Ki-67 proliferation index: correlation with prognostic parameters and outcome in multiple myeloma. Am J Clin Oncol. 2004;27:8–13. 10.1097/01.coc.0000045810.91816.41 [DOI] [PubMed] [Google Scholar]
- 38.Atrash S, Robinson M, Taneja A, Paul B, Cassetta K, Ndiaye A, et al. Bone marrow Ki-67 index is of prognostic value in newly diagnosed multiple myeloma. Eur J Haematol. 2023;111:373–81. 10.1111/ejh.14016 [DOI] [PubMed] [Google Scholar]
- 39.Møller HEH, Preiss BS, Pedersen P, Østergaard B, Frederiksen M, Abildgaard N, et al. Myc protein overexpression is a feature of progression and adverse prognosis in multiple myeloma. Eur J Haematol. 2018;101:585–90. 10.1111/ejh.13141 [DOI] [PubMed] [Google Scholar]
- 40.Li J, Zhu J, Cao B, Mao X. The mTOR signaling pathway is an emerging therapeutic target in multiple myeloma. Curr Pharm Des. 2014;20:125–35. 10.2174/13816128113199990638 [DOI] [PubMed] [Google Scholar]
- 41.Jovanović KK, Roche-Lestienne C, Ghobrial IM, Facon T, Quesnel B, Manier S. Targeting MYC in multiple myeloma. Leukemia. 2018;32:1295–306. 10.1038/s41375-018-0036-x [DOI] [PubMed] [Google Scholar]
- 42.Ramakrishnan V, Kumar S. PI3K/AKT/mTOR pathway in multiple myeloma: from basic biology to clinical promise. Leuk Lymphoma. 2018;59:2524–34. 10.1080/10428194.2017.1421760 [DOI] [PubMed] [Google Scholar]
- 43.Bieghs L, Johnsen HE, Maes K, Menu E, Van Valckenborgh E, Overgaard MT, et al. The insulin-like growth factor system in multiple myeloma: diagnostic and therapeutic potential. Oncotarget. 2016;7:48732–52. 10.18632/oncotarget.8982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hanamura I, Stewart JP, Huang Y, Zhan F, Santra M, Sawyer JR, et al. Frequent gain of chromosome band 1q21 in plasma-cell dyscrasias detected by fluorescence in situ hybridization: incidence increases from MGUS to relapsed myeloma and is related to prognosis and disease progression following tandem stem-cell transplantation. Blood. 2006;108:1724–32. 10.1182/blood-2006-03-009910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Goldschmidt H, Lokhorst HM, Mai EK, van der Holt B, Blau IW, Zweegman S, et al. Bortezomib before and after high-dose therapy in myeloma: long-term results from the phase III HOVON-65/GMMG-HD4 trial. Leukemia. 2018;32:383–90. 10.1038/leu.2017.211 [DOI] [PubMed] [Google Scholar]
- 46.Schavgoulidze A, Perrot A, Cazaubiel T, Leleu X, Montes L, Jacquet C, et al. Prognostic impact of translocation t(14;16) in multiple myeloma according to the presence of additional genetic lesions. Blood Cancer J. 2023;13:160. 10.1038/s41408-023-00933-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martello M, Poletti A, Borsi E, Solli V, Dozza L, Barbato S, et al. Clonal and subclonal TP53 molecular impairment is associated with prognosis and progression in multiple myeloma. Blood Cancer J. 2022;12:15. 10.1038/s41408-022-00610-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bladé J, Beksac M, Caers J, Jurczyszyn A, von Lilienfeld-Toal M, Moreau P, et al. Extramedullary disease in multiple myeloma: a systematic literature review. Blood Cancer J. 2022;12:45. 10.1038/s41408-022-00643-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Miyauchi M, Koya J, Arai S, Yamazaki S, Honda A, Kataoka K, et al. ADAM8 Is an Antigen of Tyrosine Kinase Inhibitor-Resistant Chronic Myeloid Leukemia Cells Identified by Patient-Derived Induced Pluripotent Stem Cells. Stem Cell Rep. 2018;10:1115–30. 10.1016/j.stemcr.2018.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Visram A, Dasari S, Anderson E, Kumar S, Kourelis TV. Relapsed multiple myeloma demonstrates distinct patterns of immune microenvironment and malignant cell-mediated immunosuppression. Blood Cancer J. 2021;11:45. 10.1038/s41408-021-00440-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jelinek T, Zihala D, Sevcikova T, Anilkumar Sithara A, Kapustova V, Sahinbegovic H, et al. Beyond the marrow: insights from comprehensive next-generation sequencing of extramedullary multiple myeloma tumors. Leukemia. 2024;38:1323–33. 10.1038/s41375-024-02206-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hochegger H, Takeda S, Hunt T. Cyclin-dependent kinases and cell-cycle transitions: does one fit all? Nat Rev Mol Cell Biol. 2008;9:910–6. 10.1038/nrm2510 [DOI] [PubMed] [Google Scholar]
- 53.Hideshima T, Nakamura N, Chauhan D, Anderson KC. Biologic sequelae of interleukin-6 induced PI3-K/Akt signaling in multiple myeloma. Oncogene. 2001;20:5991–6000. 10.1038/sj.onc.1204833 [DOI] [PubMed] [Google Scholar]
- 54.Ramakrishnan V, Kimlinger T, Haug J, Painuly U, Wellik L, Halling T, et al. Anti-Myeloma Activity of Akt Inhibition Is Linked to the Activation Status of PI3K/Akt and MEK/ERK Pathway. PloS One. 2012;7:e50005. 10.1371/journal.pone.0050005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Heredia-Guerrero SC, Evers M, Keppler S, Schwarzfischer M, Fuhr V, Rauert-Wunderlich H, et al. Functional Investigation of IGF1R Mutations in Multiple Myeloma. Cancers. 2024;16:2139. 10.3390/cancers16112139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chiang GG, Abraham RT. Phosphorylation of Mammalian Target of Rapamycin (mTOR) at Ser-2448 IsMediated by p70S6 Kinase*. J Biol Chem. 2005;280:25485–90. 10.1074/jbc.M501707200 [DOI] [PubMed] [Google Scholar]
- 57.Melick CH, Jewell JL. Regulation of mTORC1 by Upstream Stimuli. Genes. 2020;11:989. 10.3390/genes11090989 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chandrasekera P, Perfetto M, Lu C, Zhuo M, Bahudhanapati H, Li J, et al. Metalloprotease ADAM9 cleaves ephrin-B ligands and differentially regulates Wnt and mTOR signaling downstream of Akt kinase in colorectal cancer cells. J Biol Chem. 2022;298:102225. 10.1016/j.jbc.2022.102225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gunn WG, Conley A, Deininger L, Olson SD, Prockop DJ, Gregory CA. A Crosstalk Between Myeloma Cells and Marrow Stromal Cells Stimulates Production of DKK1 and Interleukin-6: A Potential Role in the Development of Lytic Bone Disease and Tumor Progression in Multiple Myeloma. Stem Cells. 2009;24:986–91. 10.1634/stemcells.2005-0220 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary methods, figures and tables S1, S5, S6
Data Availability Statement
The datasets generated and/or analyzed during the current study have been uploaded to the European Genome-Phenome Archive (EGA) (Accession: EGAS50000000392).