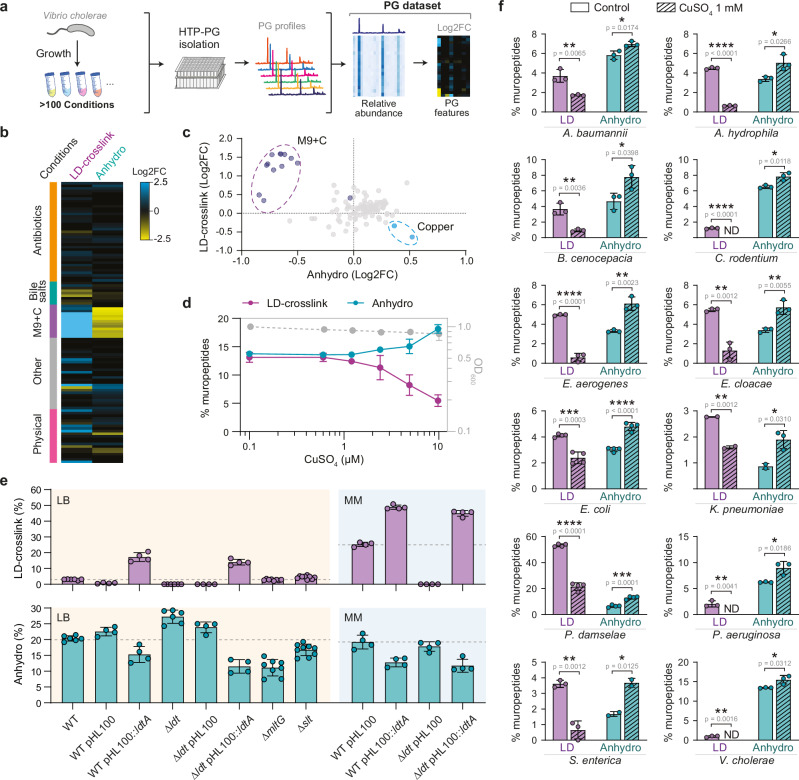
Fig. 1. Inverse correlation between LD-crosslinking and anhydromuropeptide levels.
a Overview of peptidoglycan profiling of the V. cholerae wild-type strain grown under different conditions. Peak abundances and main peptidoglycan (PG) features are calculated from the chromatographic profiles, and used for sample comparisons, studying correlations and exploration of PG dynamics. b Heatmap representing the Log2FC of LD-crosslink and anhydromuropeptide levels. The tested conditions are oriented along the y-axis, ordered by stress or the physiological condition (see Supplementary Data 1). c Scatter plot representing the Log2FC of anhydromuropeptide versus LD-crosslink levels, showing an inverse correlation between these features. PG analyses were performed in triplicate. Data are presented as mean values +/- standard deviation. M9 + C: minimal medium M9 supplemented with different carbon sources. d Concentration-dependent effect of copper on the levels of LD-crosslinking and anhydromuropeptides in the PG of V. cholerae grown in M9 medium with 0.4% (w/v) glucose (MM). e Relative levels of LD-crosslink and anhydromuropeptides in V. cholerae wild-type (WT), mutant and complemented strains grown in LB and MM. PG analyses were performed in quadruplicate (or more replicates if needed). Data are presented as mean values +/- standard deviation. f Copper inhibits LD-crosslinking and promotes anhydromuropeptide accumulation in the PG of a variety of bacterial species grown in media supplemented with 1 mM CuSO4 (Acinetobacter baumannii, Aeromonas hydrophila, Burkholderia cenocepacia, Citrobacter rodentium, Enterobacter aerogenes, Enterobacter cloacae, Escherichia coli, Klebsiella pneumoniae, Photobacterium damselae, Pseudomonas aeruginosa, Salmonella enterica and Vibrio cholerae). All bacteria were grown in LB, except for P. damselae which was grown in TSB medium. PG analyses were performed in triplicate. Data are presented as mean values +/- standard deviation. Statistical significance was determined using unpaired t-tests, with an alpha level of 0.05. Two-tailed p-values are reported. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001. ND: not detected. Source data are provided as a Source Data file.