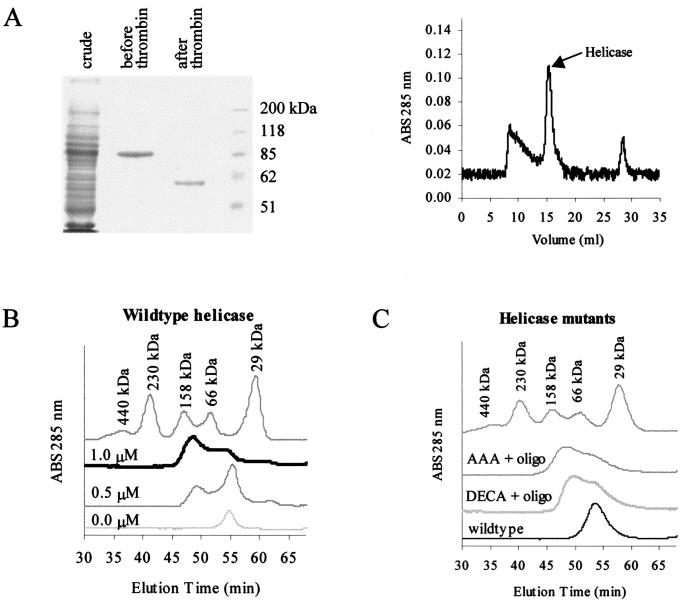
FIG. 3.
Gel filtration of the helicase protein shows that it forms a dimer in the presence of an oligonucleotide. (A) Coomassie-stained SDS-PAGE (left) of bacterially expressed GST-helicase proteins before and after thrombin cleavage shows the relative purity of this preparation. This protein thus prepared was still contaminated with other material, as shown by the absorbance graph of proteins detected after separation by FPLC (right). The fractions containing the helicase protein were collected and used for subsequent experiments. (B) In the absence of an oligonucleotide, the helicase runs at around 60 kDa, indicating that it exists as a monomer in the absence of an oligonucleotide. As increasing amounts of oligonucleotide (0, 0.5, and 1 nM) were added, the peak between the 66- and 158-kDa markers increased in height, consistent with the shift from monomer to dimer. (C) Mutants with mutations in the conserved motifs that abolish the helicase activity (DECH→DECA and TAT→AAA) can form dimers in the presence of an oligonucleotide, as shown by gel filtration. The monomeric wild-type protein without an oligonucleotide was included for comparison. ABS, absorbance.