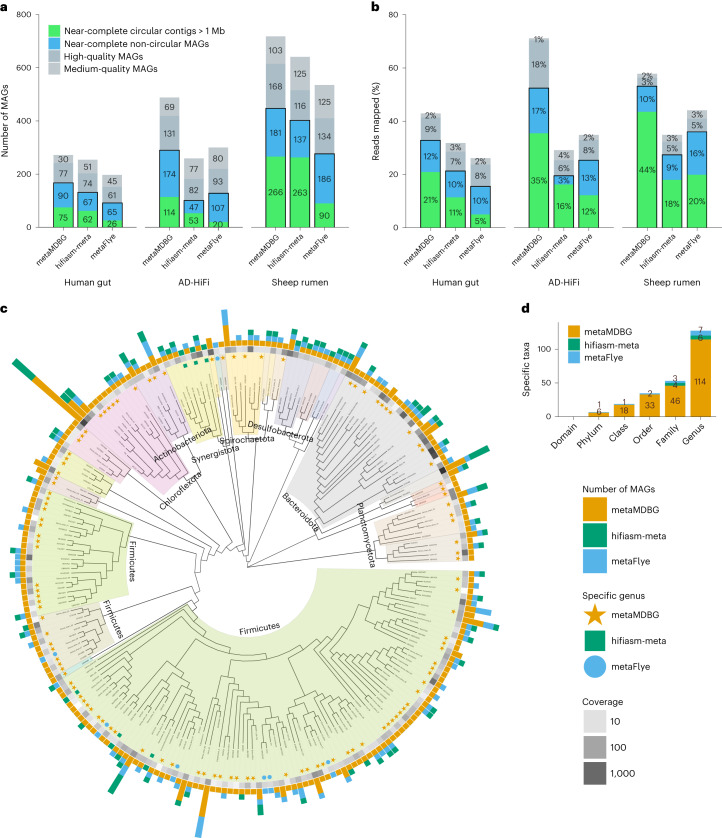
Fig. 2. Assembly results on three HiFi PacBio metagenomic projects.
a, CheckM evaluation. A MAG is considered 'near-complete' if its completeness is ≥90% and contamination is ≤5%; 'high quality' if its completeness is ≥70% and contamination is ≤10%; and 'medium quality' if its completeness is ≥50% and contamination is ≤10%. b, The percentage of mapped HiFi reads on MAGs. c, Phylogenetic tree of genera recovered from the AD-HiFi data set for all assemblers combined. For the near-complete bacterial MAGs, we generated a de novo phylogenetic tree based on GTDB-Tk marker genes, displayed at the genus level. The outer bar charts give the number of MAGs found in each genus. The colored symbols then denote genera recovered by only one of the assemblers. The grayscale heat map illustrates the aggregate abundance of dereplicated MAGs in a genus. d, Number of taxa at different levels that are unique to each assembler.