Fig. 1. LIANA+ framework overview.
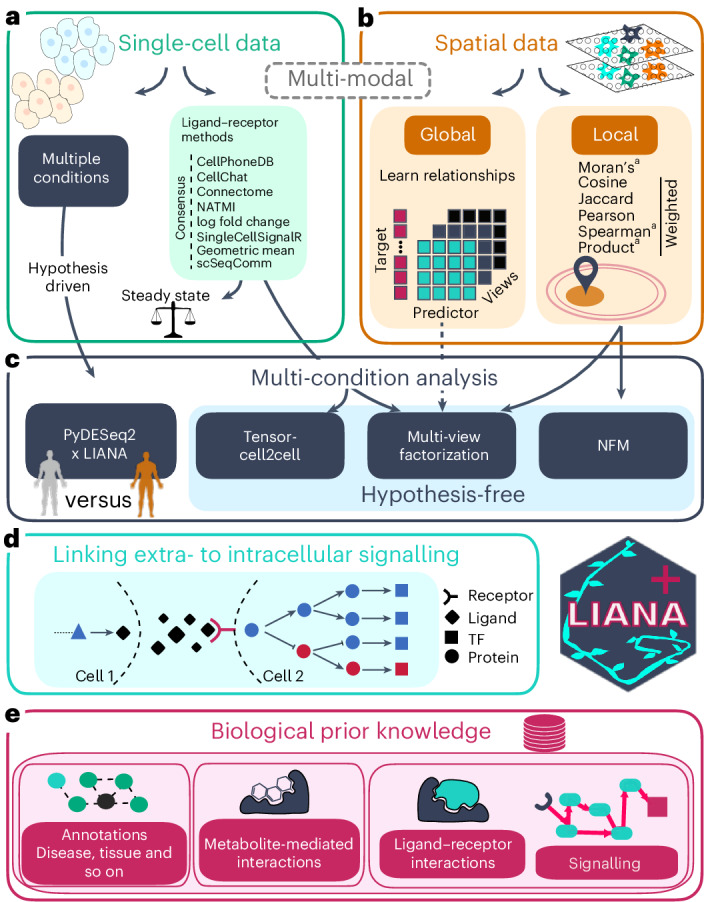
a, LIANA+ re-implements and adapts eight ligand–receptor methods to infer interactions from single-cell data, along with a flexible consensus that can integrate any combination of these methods. b, LIANA+ implements multi-view learning as well as eight local metrics to respectively capture global and local interactions from spatially resolved omics data. c, LIANA+ includes diverse strategies to identify deregulated CCC events across conditions: (1) differential CCC analysis with PyDESeq2 (refs. 47,48) for hypothesis-driven exploration and (2) unsupervised (hypothesis-free) approaches including standard NMF or higher-order factorizations via Tensor-cell2cell20 and multi-view factor analysis43. d, LIANA+ connects intercellular interactions to intracellular signalling pathways using sign-coherent network optimization. e, LIANA+ is built on a rich knowledge base—OmniPath22 and BioCypher59—which comprise ligand–receptor interactions and annotations, including such mediated by metabolites68, as well as intracellular knowledge, such as signalling pathways and TFs. Finally, all components (a–e) of LIANA+ are applicable to both dissociated single-cell and spatially resolved multi-omics data. aFor spatially weighted Spearman correlation, along with the standard metric, we implemented its masked version from scHOT32; for Moran’s R we adapted both the global and local versions from SpatialDM8 and for the spatially weighted product, we also included a max-normalized version.
