Fig. 2. LIANA+ uses standardized inputs and outputs to streamline the inference intercellular and intracellular signalling.
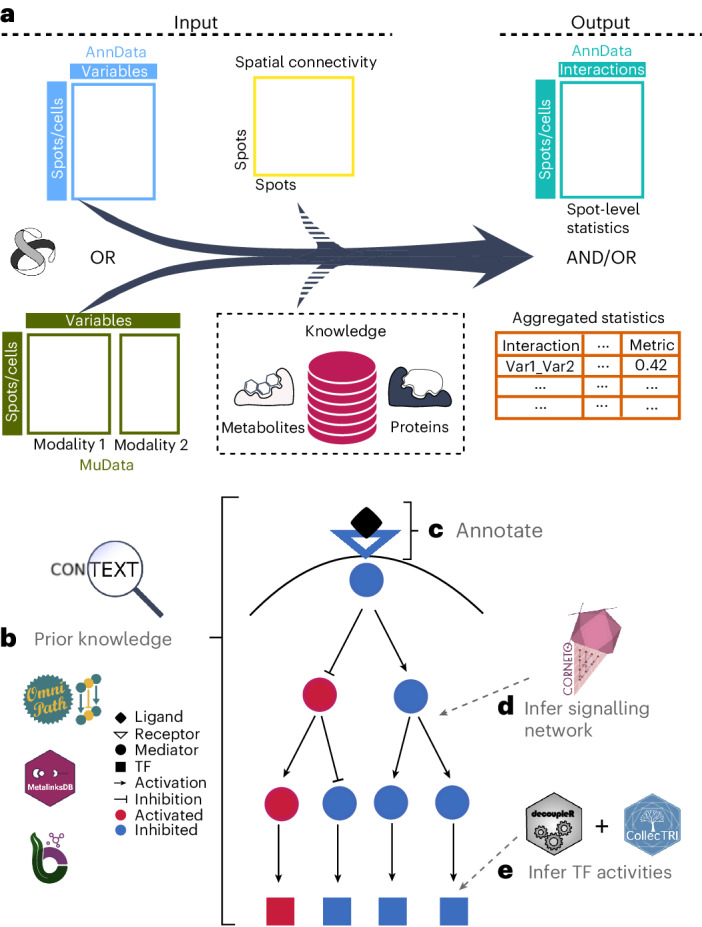
a, LIANA+ accepts unimodal (AnnData) or multi-modal (MuData) data objects as inputs with (optional) prior knowledge and/or spatial information. These are then transformed into data frames with aggregated interaction results or unimodal objects with statistics at the individual spot or cell level. To enable the inference of CCC across modalities, the methods implemented in LIANA+ accept MuData objects77 as input. These provide essential functionalities to load and store multi-modal data77, and can be thought of as an extension of AnnData objects78, which are the default input of LIANA+ when working with unimodal single-cell or spatial data. b,c, LIANA+ makes use of existing prior knowledge frameworks (b)22,59,68 to annotate interactions according to, for example, pathways, disease or location (c). d, Similarly, it uses this prior knowledge to infer putative causal (sign-coherent) signalling networks49,79, emanating from ligand–receptor interactions down to active TFs. e, The TF activities can be estimated by making use of generalistic regulon prior knowledge80 and standard enrichment analyses81.
