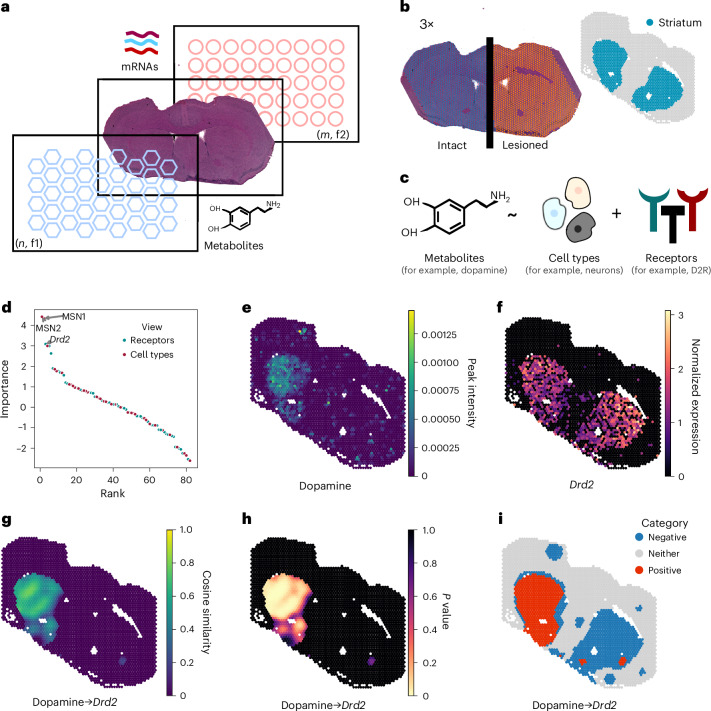
Fig. 3. LIANA+ models intercellular communication from spatial multi-omics data.
a, Spatially resolved transcriptomics (10X Visium) and metabolomics (matrix-assisted laser desorption/ionization mass spectrometry imaging) yield two matrices with different sets of features for each modality (f1 or f2), and observations (n or m) which correspond to different locations captured on the same tissue section26. b, Parkinson’s disease mouse model annotated for striatum in intact and lesioned hemispheres, with three replicates (3✕). c, Multi-view modelling integrates metabolite peak intensities, brain-specific receptor expression and cell-type proportions to identify their spatial relationships. This approach enables the estimation of joint performance and individual contributions of receptor expression and cell-type proportions in predicting metabolite peak intensities. Cell-type proportions were deconvoluted35 using a murine single-cell atlas36 as a reference. d, Dopamine predictors ranked according to their median importance (y axis; ordinary least squares t-values), with names shown for the top three predictors: Drd2 and MSNs 1 and 2. e, Normalized dopamine peak intensities. f, log1p Drd2 receptor gene expression. g–i, Local interactions between dopamine and its canonical D2R receptor, encoded by Drd2, as measured by spatially weighted cosine similarity (g), its corresponding uncorrected permutation P values (h) and interaction categories (i). The images showcase slide B1 from experiment V11L12-109 (ref. 36). Source numerical data are available in source data.