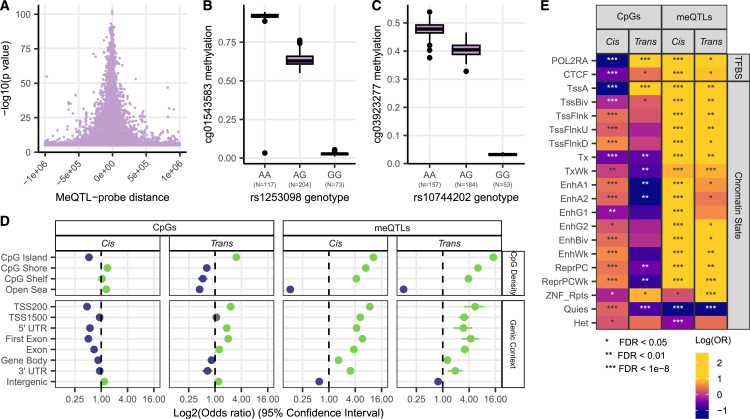
Figure 2.
Genetic effects on the whole-skin methylome
(A) Distance between lead cis-meQTL and CpG vs. strength of association of the meQTL effect.
(B) Most associated cis-meQTL association between variant rs1253098 (chr14:59945536) and cg01543583 (chr14:59947673).
(C) Most associated trans-meQTL association between the variant rs10744202 (chr12:125800244) and cg03923277 (chr12:104359732).
(D) Enrichment and depletion of meQTL effects in different genomic contexts. Green indicates significant (FDR 0.05) enrichment, purple indicates significant depletion. CpG islands based on UCSC refGene annotations (CpG shore, 2 kbp from CpG island; CpG shelf, 2–4 kbp from CpG island; open sea, 4 kbp from CpG island), and gene context is based on refGene annotations (TSS200, 200 bp from transcription start site (TSS); TSS1500, 200–1,500 bp from TSS).
(E) Enrichment and depletion of meQTL effects in transcription factor binding sites (TFBSs) in suprapubic skin and genomic states in leg skin generated using an 18-state ChromHMM model from the Roadmap Epigenomics Consortium.59 White asterisks indicate significant depletion, black asterisks indicate significant enrichment. Suprapubic skin TFBS and leg skin chromatin state annotations were obtained from the EpiMap Repository60 (TssA, active TSS; TssBiv, bivalent/poised TSS; TssFlnk, flanking active TSS; TssFlnkU, flanking TSS upstream; TssFlnkD, flanking TSS downstream; Tx, strong transcription; TxWk, weak transcription; EnhA1, active enhancers 1; EnhA2, active enhancers 2; EnhG1, genic enhancers 1; EnhG2, genic enhancers 2; EnhBiv, bivalent enhancer; EnhWk, weak enhancer; ReprPC, repressed polycomb; ReprPCWk, weak repressed polycomb; ZNF_Rpts, ZNF genes and repeats; Quies, quiscent chromatin; Het, heterochromatin; see Kundaje et al.59).