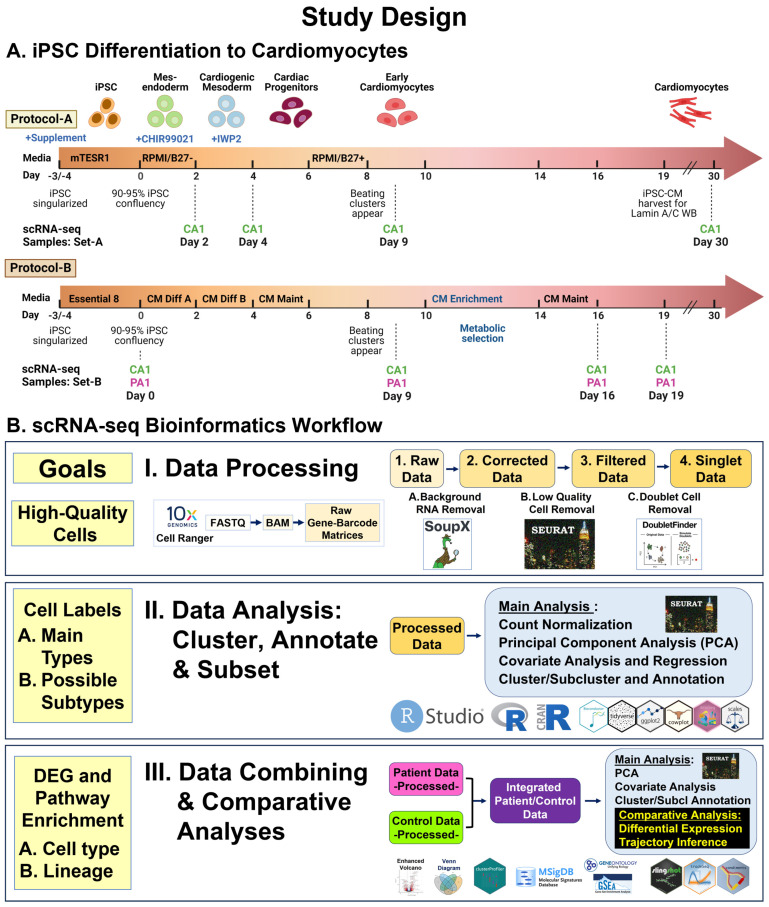
Figure 1.
Study design. (A) iPSC differentiation into cardiomyocytes. Using standard protocols, Protocol-A and Protocol-B, we differentiated three iPSC lines (CA1-A, CA1-B, PA1) and collected 12 cell samples (8 from CA1 and 4 from PA1), across seven time points, for scRNA-seq. Using Protocol-A, we differentiated six iPSC lines (CA1-B, U2, CA3, PA1, PA2, PA3) and collected six cell samples (3 from controls and 3 from patients) at Day 19 for Western blot analysis. (B) scRNA-seq bioinformatics workflow. We used standard pipelines and software packages for data processing and analyses. Three main steps were involved: I. data processing for initial read processing/mapping and sequential quality control processing of raw data sets; II. data analysis for identification of main cell types and possible cell subtypes by unsupervised clustering, annotation, and subset analyses; III. data combining and comparative analyses of samples (Patient vs. Control) for cell type and lineage differentially expressed genes (DEGs) and gene set enrichment.