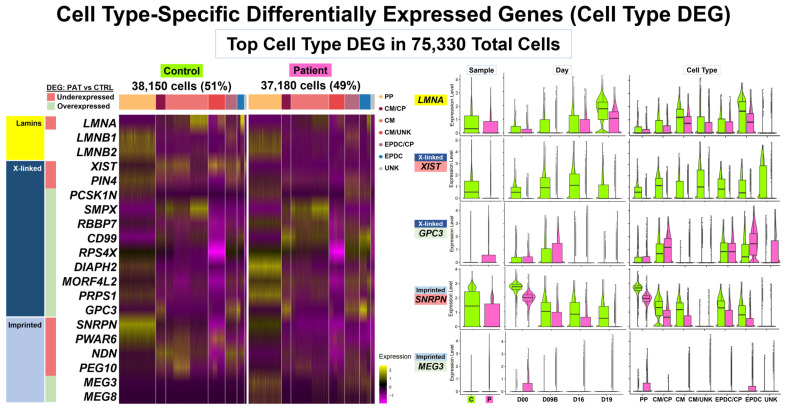
Figure 3.
Cell type-specific differentially expressed genes (cell type DEGs): top DEGs. Our comparative analyses (Patient vs. Control cells) included cell type-specific differential expression and enrichment using integrated data at four time points: D00, D09B, D16, and D19. We identified cell type DEGs in six pairs of ‘balanced’ cell types/subtypes, with 75,330 total cells. Left panel: heat map for top DEGs in main cell types for 38,150 (51%) Control cells and 37,180 (49%) Patient cells. Top DEGs included LMNA, 11 X-linked (XL) genes, and six imprinted genes. Right panel: violin plots with boxplot by sample, day, and cell type for LMNA, XL genes, XIST and GPC3, and imprinted genes, SNRPN and MEG3. Overall, these results show Patient compared to Control cells had LMNA underexpressed in both CMs and EPDCs at Day 19, limited XIST expression for all time points and shared cell types, XL gene GPC3 overexpressed in CM/CP and EPDCs, SNRPN underexpressed for all time points and most shared cell types, and MEG3 overexpressed in PPs and EPDCs.